-
Paper Information
- Previous Paper
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
International Journal of Modern Botany
p-ISSN: 2166-5206 e-ISSN: 2166-5214
2013; 3(2A): 34-42
doi:10.5923/s.ijmb.201310.05
The Genetic Basis of Overcompensation in Plants: A Synthesis
Daniel R. Scholes 1, 2, Madhura H. Siddappaji 1, 3, Ken N. Paige 1, 2
1School of Integrative Biology, University of Illinois, Urbana, Illinois 61801
2Program in Ecology, Evolution, and Conservation Biology, University of Illinois, Urbana, Illinois 61801
3Department of Biological Science, 319 Galvin Life Science, University of Notre Dame, Notre Dame 46656
Correspondence to: Ken N. Paige , School of Integrative Biology, University of Illinois, Urbana, Illinois 61801.
| Email: |  |
Copyright © 2012 Scientific & Academic Publishing. All Rights Reserved.
Although genetic variation for compensation exists, studies of the mechanisms underlying such variation have only recently been reported. Here we synthesize, and add new information, on what is known to date about the genetic underpinnings of plant compensation following insect and mammalian herbivory with a focus predominantly on the phenomenon of overcompensation. These studies have centered on the role of endoreduplication in compensation and genes involved in the compensatory response. Specifically, we propose that plants with the capability of overcompensating (increasing both biomass and fitness when compared to undamaged controls) reprogram their transcriptional activity in at least three important ways: 1) through a suite of defensive mechanisms, 2) through an increase in expression of genes involved in energy metabolism and 3) through an increase in DNA content (via endoreduplication), with the increase in DNA content feeding back on pathways involved in defense and metabolism through increased gene expression.
Keywords: Endoreduplication, Glucose-6-Phosphate-1-Dehydrogenase, Overcompensation
Cite this paper: Daniel R. Scholes , Madhura H. Siddappaji , Ken N. Paige , The Genetic Basis of Overcompensation in Plants: A Synthesis, International Journal of Modern Botany, Vol. 3 No. 2A, 2013, pp. 34-42. doi: 10.5923/s.ijmb.201310.05.
Article Outline
1. Introduction
- Plants have evolved a variety of mechanisms for reducing the negative effects of herbivory, including the production of structural and chemical defensive traits that reduce or prevent tissue damage by herbivores and tolerance strategies that allow plants to compensate for tissues lost with little or no decrement in fitness[11, 32]. In particular, interest in tolerance was motivated by empirical studies demonstrating that herbivore damage can, under certain circumstances, increase, rather than decrease, plant reproductive success (a specialized case termed overcompensation, i.e., increased flower, fruit, and seed production following herbivory) [32]. Specifically, Paige and Whitham[23] showed that when mule deer and elk removed 95% or more of the aboveground biomass of the monocarpic biennial scarlet gilia, Ipomopsis aggregata, the product of lifetime seed production, seed germination, and seedling survival averaged 3.0 times that of uneaten controls[2, 20, 21, 22]. Evidence for increased flower, fruit and seed production following herbivory has also been found for numerous plant species since the initial study of Paige and Whitham[23] including Ipomopsis arizonica[16], Gentianella campestris, G. amarella[14, 19], Arabidopsis thaliana[17, 37], Erysimum strictum[26] and Solanum tuberosum [25] to name but a few. Evidence that genetic variation for compensation also exists. Specifically, some families exhibit overcompensation, whereas others express only patterns of equal- or undercompensation[10, 14, 17, 28, 34, 37]. Although these observations provide evidence that genetic variation for compensation exists, studies of the mechanisms underlying such variation have only recently been reported. Here, we review and add new information on what is known to date about the genetic underpinnings of plant compensation following insect and mammalian herbivory with a focus predominantly on the phenomenon of overcompensation. These studies have centered on the role of endoreduplication in compensation and genes involved in the compensatory response.
2. Role of Endoreduplication in Compensation
2.1. Fitness Compensation and Chromosomal Plasticity
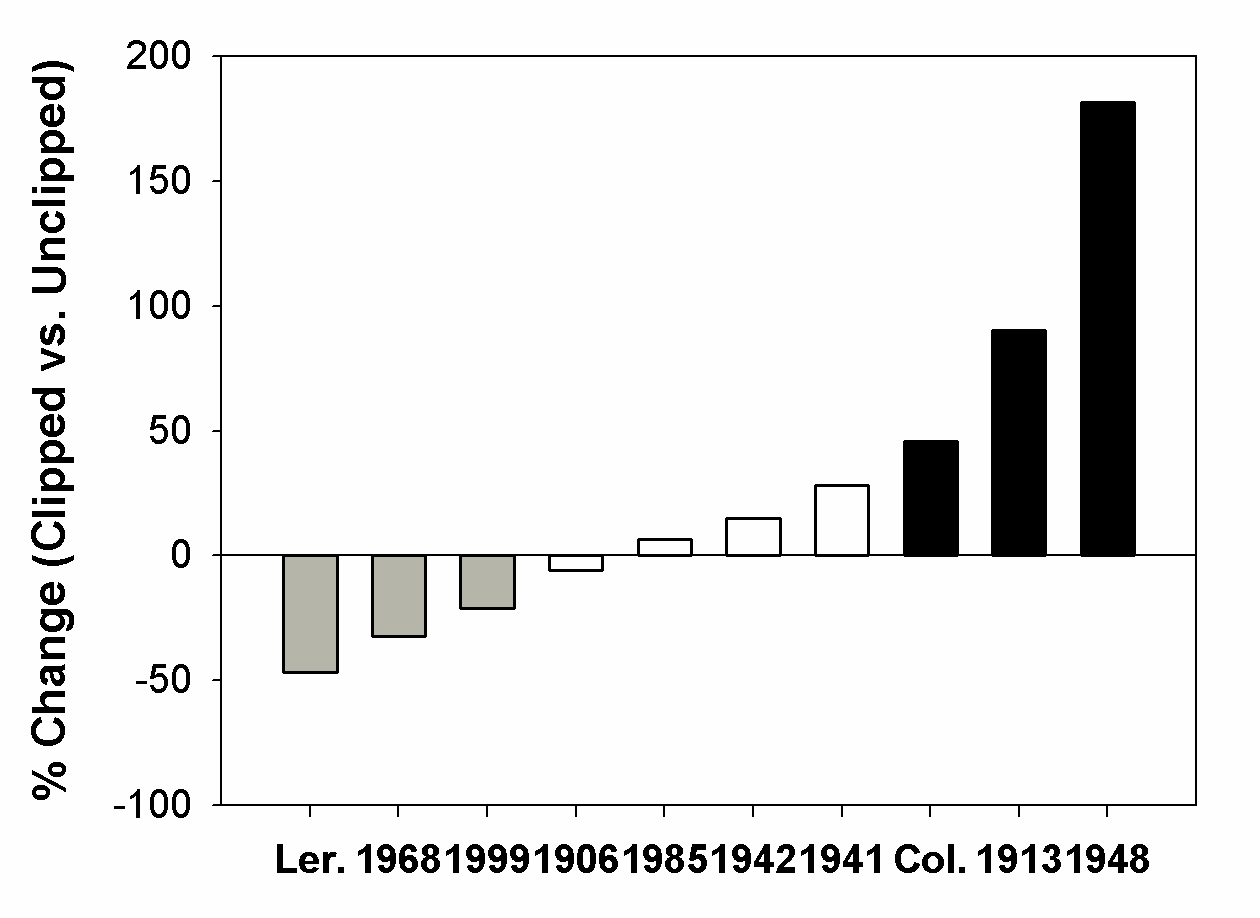
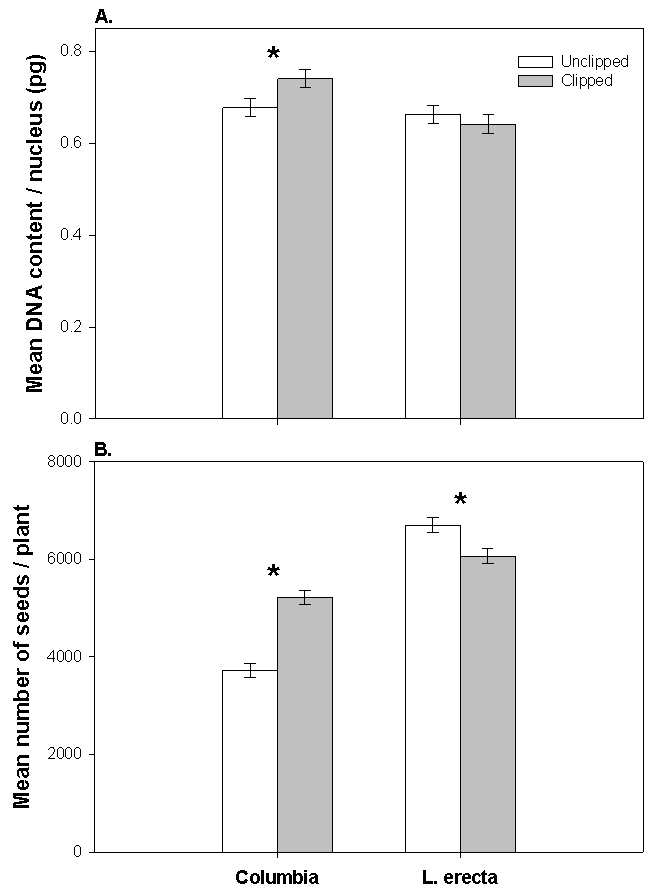
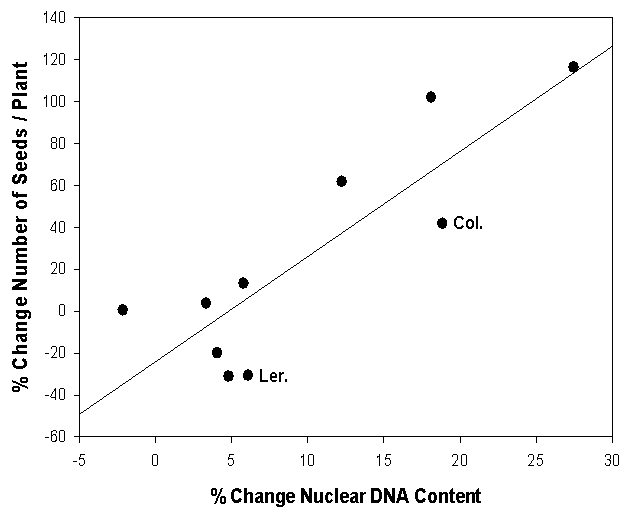
- Over the past several years we have been testing the validity of a novel idea - that endoreduplication leads to enhanced growth and reproduction following herbivory, explaining the phenomenon of tolerance and overcompensation in plants[28]. Endoreduplication is the replication of the genome without mitosis, which leads to endopolyploidy, an increase in cellular chromosome number. This process is common in many groups of eukaryotes and angiosperms[3, 18, 33]. What is unknown is the degree to which endoreduplication is genetically/genotypically plastic following herbivory. The removal of apical dominance reduces the level of auxin, leading to axillary bud break and stem regeneration - high levels of auxin are also known to repress the endocycle and by contrast, lower levels of auxin trigger an exit from mitotic cycles and an entry into endocycles[9]. Thus, there is a direct link between endoreduplication and the removal of apical dominance (a common form of herbivory by both mammals and insects; e.g.,[2]). It is also important to point out that endoreduplication occurs in cells within the stem tissue but does not occur in reproductive cells[9]. Ecotypes and recombinant inbred lines of Arabidopsis thaliana have different compensatory capabilities following the removal of apical dominance[28, 31]. Removal of approximately 85% of the primary inflorescence (leaving 1cm of stem, simulating natural mammalian herbivory) of 92 recombinant inbred lines (RILs) of Arabidopsis (developed from a cross between Columbia and Landsberg erecta ecotypes; Lister and Dean[15] show a range of compensatory responses from undercompensation to overcompensation for silique and seed production following natural herbivory[37] and experimental clipping[31]. Among these, 12 lines overcompensated, 9 undercompensated and 71 equally compensated (i.e., significantly more, fewer or equal numbers of siliques/plant, respectively; p<0.05) [see 31 for details of our statistical analysis]. The parental types represent a subset of extreme ecotypes of the compensatory continuum, with RILs ranging from undercompensation (e.g., CS1968 with a 32% reduction in number of siliques/plant) to overcompensation (e.g., CS1948 with 181% increase in number of siliques/plant, Figure 1) following apical damage.Different ecotypes of Arabidopsis thaliana also differ in the degree of endopolyploidy following the removal of apical dominance, and the degree of endopolyploidy achieved is positively correlated with measures of fitness[28]. Clipped plants of Columbia have a significantly lower proportion of nuclei at the basal 2C ploidy level, and thus a higher proportion of nuclei at endopolyploid levels (4C, 8C, and 16C) when compared to unclipped controls. In contrast, clipped plants of Landsberg erecta showed no significant differences in endopolyploidy relative to unclipped plants. These results indicate an increase in overall DNA content per cell in Columbia when clipped, while clipped plants of Landsberg erecta show no significant difference in the amount of DNA per cell (Figure 2A). It is also important to point out that base-line ploidy of clipped rosettes (prior to bolting) did not differ from unclipped rosettes for either Columbia or Landsberg erecta (P<0.15). Given that only inflorescence tissue was removed during clipping, rosettes were not expected to differ between treatments within an ecotype and may serve as verification that changes in ploidy in the inflorescences were due to the clipping treatment. Additionally, clipped plants of Columbia produced significantly greater numbers of siliques and seeds (51% and 40% more, respectively) than plants that were not clipped while clipped plants of Landsberg erecta produced an equal number of siliques and fewer seeds when compared to those that were not clipped (Figure 2B for number of seed produced). Above-ground biomass was significantly greater for both clipped plants of Columbia and Landsberg erecta when compared to their unclipped controls, although the magnitude of increase was greater for Columbia (49% versus 24% more, respectively). There was also a significant genotype X environment (clipping) interaction between Landsberg erecta and Columbia for endopolyploidy (P=0.008).
2.2. Endoreduplication Contributes to Rapid Regrowth and Attributes of Fitness
- Increasing chromosome number, and thus gene copy number, may provide a means of increasing gene expression, likely by the up-regulation of selected genes or gene families (transcriptomic studies indicate that only a subset of genes are differentially regulated;[31]). Furthermore, increasing chromosome number increases the total DNA content and hence cell size leading to extensive cell growth through endoreduplication. Growth by cell division along with growth by cell expansion through endoreduplication may be faster than growth by cell division alone[3]. Also, rapid growth and development following the removal of apical dominance may be further enhanced by an increase in cell size by maximizing nutrient transport, protein synthesis, and light and water absorption[13]. Endoreduplication also occurs predominantly among plants adapted to habitats that require fast growth and development[3, 4]. This correlation between life history and endoreduplication suggests a possible functional relationship between enhanced chromosome production and rapid regrowth following the removal of apical dominance[28]. There are also direct ties between endoreduplication and seed development. Endoreduplication occurs widely in suspensor cells, which connect the embryo to the surrounding nourishing tissue and ensures nutrient transfer; plasmodesmata of the cell walls connecting the cytoplasm of adjacent cells slows down nutrient transport, thus, faster transport is facilitated by producing larger and fewer cells with fewer cell walls via endoreduplication[3]. Kowles and Phillips[12] suggested that extra DNA produced by endoreduplication is important in maize endosperm development and kernel filling through increased gene expression and protein synthesis. However, high temperatures or water deficits can cause the endosperm of developing seeds to remain primarily mitotic, reducing endoreduplication leading to smaller endosperm that are ill suited in supporting the embryo[6, 13].
2.3. Endoreduplication is Genetically Regulated and can be Experimentally Manipulated
- The endocycle consists of alternating G and S phases without mitotis or cytokinesis and is considered an alternative to the typical cell cycle (with phases G1, S, G2, and M), largely due to the wide range of gene families that are common to the regulation of both cycles. Specifically, there are numerous genes that regulate various aspects of endoreduplication and the endocycle, including the time of endocycle induction and the number of endocycles completed[8, 35]. One such gene family, the cyclin A2 (CYCA2) class of cyclins, is involved in the checkpoint between the G phase and the S phase of cells that have entered the endocycle. This class of cyclins activates a cyclin-dependent kinase (CDK1) that represses cell entry into the S phase (DNA replication phase). Regulation of CYCA2 proteins, therefore, would not affect the time of induction or the number or types of cells entering the endocycle, but it would affect the number of successive rounds of DNA replication via endoreduplication. Increased Level of Polyploidy1 (ILP1) has been shown to be a transcriptional repressor of the CYCA2 family of cyclins[38], where reducing ILP1 expression by T-DNA insertion in the fifth intron of ILP1 (such that transcripts are truncated) of A. thaliana Columbia plants caused a significant increase in CYCA2 family transcription and a significant decrease in endoreduplication compared to wildtype Columbia. Likewise, overexpression of ILP1 via CaMV 35S promoter caused a significant decrease in CYCA2 family transcription and a significant increase in endoreduplication compared to wildtype Columbia[38]. Due to the demonstrated control over the endocycle, ILP1 is an appropriate target for manipulating the degree of endopolyploidy. Specifically, we hypothesize that endoreduplication plays a direct role in the compensatory ability of A. thaliana to herbivory, such that an increase in the number of DNA synthesis cycles in endoreduplicating cells will result in higher fitness (biomass, number of flowers, siliques, and seeds) following simulated herbivory than unclipped controls, and that a decrease in the number of DNA synthesis cycles in endoreduplicating cells will result in lower fitness following simulated herbivory than unclipped controls. By experimentally manipulating the degree of endopolyploidy achieved in the plant and then measuring plant fitness, we can investigate directly the potential adaptive significance of endoreduplication. We are currently using ILP1 overexpression and knockdown lines along with the Columbia parental line as a control. These results will be reported elsewhere.
|
3. Genes Involved in Compensation
- Although we know a great deal about the genetic basis of endoreduplication per se[8, 35, 38] and have evidence that it plays a role in fitness compensation[28] (Scholes and Paige, unpublished data) we are just beginning to uncover the genes and gene pathways affecting fitness compensation following endoreduplication. In a recent study, combining microarray and QTL analyses, we uncovered one gene that appears to play a significant role in overcompensation;glucose-6-phosphate-1-dehydrogenase[31].
3.1. QTL/Microarray Analyses
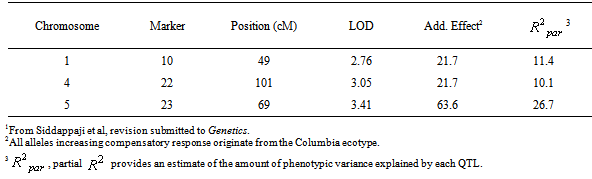
- From the QTL mapping studies using the Lister and Dean lines[15], showing the full range of compensatory responses from under to overcompensation, we uncovered three QTL explaining 11.4%, 10.1% and 26.7% of the variation in fitness compensation (Table 1,[31]). These three QTL showed additive effects increasing silique production by an average of 21.7 to 63.6 siliques upon clipping. All alleles increasing fitness were contributed by the Columbia ecotype (Table 1), although the compensatory response distribution suggests contributions from Landsberg erecta (i.e., evidence for transgressive segregation, see Figure 1).Although QTL can help in identifying regions of the genome responsible for compensation, it is difficult to identify specific candidate genes, as a single QTL likely contains hundreds of genes (a single QTL ranges from10-20cM in size with ~1cM of Arabidopsis covering 210kb of the genome[24]) of which some may and some may not be responsible for observed patterns of fitness compensation. Considering the number of QTL obtained, we combined QTL mapping with microarray expression data of clipped and unclipped plants of the Columbia ecotype to help in identifying potential candidate genes. Wayne and McIntyre[36], for example, successfully combined data from QTL and microarrays to identify genes responsible for ovariole number in Drosophila melanogaster. From our microarray analysis a total of 109 genes were found to be differentially expressed between clipped and unclipped plants of Columbia. Of these, 30, 19, 17, 16 and 27 differentially expressed genes were located on chromosomes 1 through 5, respectively between clipped and unclipped plants. Based on a gene ontology analysis, these genes can be generally classified into stress response genes, metabolic genes, and growth/reproductive genes. When mapped with the QTL data, only a single gene co-localizes within one of the QTL markers (QTL 3 located on chromosome 5, at 69 cM, Table 1), a glucose-6-phosphate-1-dehydrogenase (G6PDH1: EC 1.1.1.49;[31]).
3.2. G6PDH1: T-DNA Knockout and Complementation Studies
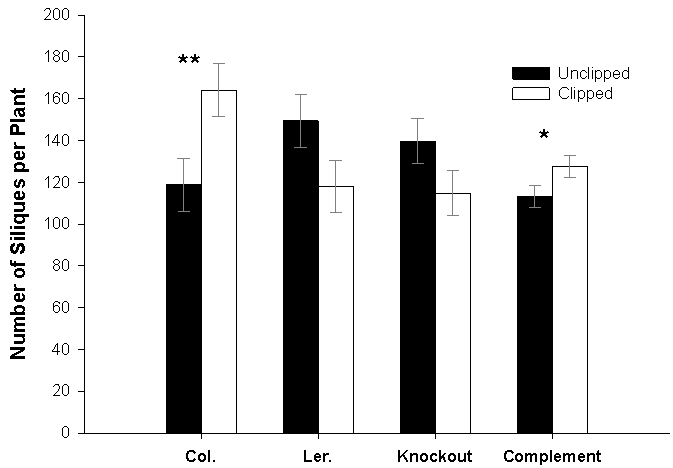
- To assess the role of G6PDH1, T-DNA gene knockout experiments and a complementation study, wherein G6PDH1 was reintroduced into a T-DNA gene knockout line. Knockout studies of T-DNA insertion lines of G6PDH1 (sharing the same genetic background as Columbia) showed patterns of equal compensation, with a trend toward undercompensation, rather than overcompensation as observed in the Columbia wild-type (see Figure 4 for an example). Furthermore, G6PDH1 expression data through time comparing Landsberg erecta, an undercompensating ecotype, and Columbia, an overcompensating ecotype, showed higher levels of expression (1.4- to 2.2-fold) in Columbia following the removal of apical dominance, data consistent with our knockout experiments wherein lowering or knocking out G6PDH1 expression resulted in equal to undercompensation instead of overcompensation in Columbia.Our transgenic line complemented with G6PDH1 restored the compensatory response from equal compensation, with a trend toward undercompensation, in the knockout line to overcompensation (at p = 0.064)[31]. We suspect that positional effects of the transgene or unmeasured environmental influences may have constrained the magnitude of the compensatory response typically observed in Columbia. Nonetheless, these results support the importance of G6PDH1 in regulating the compensatory response following the removal of apical dominance.
3.3. SNF1-related Kinases
- Using 11C photosynthate labeling as well as sugar and enzyme measurements following simulated insect leaf herbivory (wounding and regurgitant from Manduca sexta) on Nicotiana attenuata, Schwachtje et al.[30] found rapid changes in sink-source relations that increased the allocation of sugars to roots. This herbivore-induced response is regulated by the β-subunit of a SnRK1 (SNF1-related kinase) protein kinase, GAL83, which increases assimilate transport to roots. The SNF1-related kinase was identified by mRNA differential display[7]. If herbivory occurs early in development the plant is able to enhance root reserves and delay senescence, prolonging the period of reproduction and hence better tolerate herbivory.
4. Mechanisms of Compensation
- Although we use the term tolerance in reference to the suite of compensatory responses in this article, we think it is noteworthy to point out that not all interactions between plants and herbivores are necessarily antagonistic as indicative of the use of the term “tolerance” – making the best of a bad situation by mitigating the effects of damage. There is some evidence, albeit limited, suggesting that some plant populations have adapted to being eaten given that plant fitness is maximized following herbivory[1], i.e., in some cases of overcompensation. For example, studies comparing historically grazed and ungrazed populations of the plant Gentianella campestris indicate that repeatedly grazed populations overcompensate, while ungrazed populations remain completely intolerant[14]. Nonetheless, studies on endoreduplication and molecular genetics suggest that mechanistically compensation represents a continuum of differential regulatory responses caused by genetic differences within the same pathways. For example, there is considerable sequence variation in G6PDH1, with three non-synonymous substitutions, each causing a change in an amino acid, between Columbia and Landsberg erecta that may explain the differential patterns of expression in G6PDH1 following apical damage and regrowth and perhaps the differences in compensation (Max Planck Institute for Developmental Biology, POLYMORPH Project, http://polymorph-clark20.weigelworld.org/cgi-bin/retrieve_cds_snp.cgi).We propose that plants with the capability of overcompensating (increasing both biomass and fitness when compared to undamaged controls) reprogram their transcriptional activity in at least three important ways: 1) through a suite of defensive mechanisms, 2) through an increase in expression of genes involved in energy metabolism and 3) through an increase in DNA content (via endoreduplication,[28]), with the increase in DNA content feeding back on pathways involved in defense and metabolism through increased gene expression[31]. Initially, following apical damage, a suite of defensive reactions are elicited from the cellular damage associated with herbivory. These may include reactive oxygen species to ward off infection and induced chemical defenses, such as glucosinolates, via the shikimate pathway[27]. When analyzing genes that were significantly differentially expressed (from our microarray data), several of the genes affected were found to be enzymes (e.g., a suite of invertase genes, G6PDH1, and galactinol synthase) involved in carbohydrate metabolism and these genes were significantly up-regulated and likely play a significant role in overcoming tissue loss. In addition, up-regulation of G6PDH1 ultimately leads to the biosynthesis of nucleic acids (all part of the oxidative pentose-phosphate pathway), consistent with the significant increase in DNA content (through endoreduplication) observed in overcompensating ecotypes of Arabidopsis thaliana when compared to undercompensating ecotypes[28, 31]. Interestingly, removal of apical dominance reduces the level of auxin leading to axillary bud break and stem regeneration and low levels of auxin trigger an exit from mitotic cycles into the endocycle[9]. Thus, there is a direct link between endoreduplication and the removal of apical dominance. Schwachtje et al.[30] found rapid changes in sink-source relations that increased the allocation of sugars to roots using simulated insect herbivory. This herbivore-induced response regulated by the β-subunit of a SnRK1 (SNF1-related kinase) protein kinase increases assimilate transport to roots ultimately enhancing root reserves (and perhaps the down-regulation of systemin –[29]). Schwachtje et al.[30] also found an increase in vacuolar invertase activity in the roots of simulated insect damaged plants. Invertases feed glucose into the OPP pathway in which G6PDH1 resides[5]. Based on promoter-reporter fusion studies (G6PDH1 promoter: β-glucuronidase (GUS)) and subsequent histochemical staining G6PDH1 is expressed in virtually all tissues rather than localized to any specific tissue (Siddappaji and Paige, unpublished results). As noted above, up-regulation of G6PDH1 ultimately leads to the biosynthesis of nucleic acids, consistent with the significant increase in DNA content (through endoreduplication;[28]), with the increase in DNA content leading to greater metabolism through increased gene expression. These results are consistent with patterns of regrowth observed following clipping in Arabidopsis. Increased expression of a variety of invertases and G6PDH1 in the overcompensating genotype Columbia appears to facilitate the rapid regrowth, increased biomass, and ultimately enhanced fitness following the removal of apical dominance, as evidenced by plant-wide localization of G6PDH1 (Siddappaji and Paige, unpublished results). Interestingly, root biomass is also enhanced in Ipomopsis aggregata following ungulate herbivory in which 95% of the aboveground biomass is removed, apparently creating a sink for carbon and ultimately resulting in enhanced fitness[23]. A SNF1-related kinase and invertases may be involved in transporting and enhancing carbon in the roots of I. aggregata (and perhaps Arabidopsis) as well following mammalian herbivory, ultimately leading to enhanced fitness. Overall, we suggest that these results represent only a snapshot in time of the molecular biology of compensatory responses following herbivory and that there is much to uncover. Future studies will clearly need to track molecular genetic changes through time to understand the dynamic variation of the compensatory response and the pathways involved.
ACKNOWLEDGEMENTS
- Research was funded by NSF grants DEB-0522409, DEB-1010868, and DEB-1146085 to K.N. Paige. We wish to acknowledge two anonymous reviewers for their thoughtful suggestions that have helped to improve this manuscript.
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML