-
Paper Information
- Next Paper
- Previous Paper
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
International Journal of Biophysics
p-ISSN: 2168-4979 e-ISSN: 2168-4987
2013; 3(1A): 9-17
doi:10.5923/s.biophysics.201311.02
Introduction of FRET Application to Biological Single-molecule Experiments
Kenji Okamoto
Cellular Informatics Laboratory, RIKEN, Wako, Saitama, 351-0198, Japan
Correspondence to: Kenji Okamoto, Cellular Informatics Laboratory, RIKEN, Wako, Saitama, 351-0198, Japan.
| Email: |  |
Copyright © 2012 Scientific & Academic Publishing. All Rights Reserved.
Since the fluorescence resonance energy transfer (FRET) measurement is based on the fluorescence measurement technology, the signal can be obtained from single molecules. The single-molecule FRET (smFRET) measurement has already been extensively used in the field of biology. The combination of the single-molecule sensitivity, the nanometer-scale spatial resolution and the realtimeness gives us the exclusive information on the structure and the dynamics of biomolecules, such as coexisting multiple conformational states or temporal evolution of conformational changes. As an introductory review of the smFRET measurement, this article briefly explains the theory, the basic apparatus, the typical data analysis methods and some examples of experiments applied to biomolecules.
Keywords: FRET, Single-molecule Measurement, Molecular Structure, Conformational Dynamics
Cite this paper: Kenji Okamoto, Introduction of FRET Application to Biological Single-molecule Experiments, International Journal of Biophysics , Vol. 3 No. 1A, 2013, pp. 9-17. doi: 10.5923/s.biophysics.201311.02.
Article Outline
1. Introduction
- The fluorescence resonance energy transfer (FRET) has been extensively used in biological experiments. According to its nature that the distance between two fluorophores can be detected as the ratio of fluorescence intensities, it is sometimes referred to as a “spectroscopic ruler”[1] and is often used to examine the molecular structure. Since the FRET measurement is typically most sensitive at the distance of several nanometers, which is comparable to the dimension of biomolecules or their structural dynamics, its application to biological molecules seems to be inevitable. In addition, since the FRET measurement is based on the fluorescence measurement technology, it can be carried out with the single molecule sensitivity.FRET was experimentally applied to biological molecules after a while from its formulation by Förster[2]. The dependence on the distance[3] and the relative orientation[4] between fluorophores was shown to be consistent with the Förster’s theory. On the other hand, advances in fluorescence microscopy enabled the single- molecule measurement, so-called the single-molecule detection (SMD) or the single-molecule spectroscopy (SMS), in 1990s. Single fluorescent molecules on the glass surface were successfully imaged by using the scanning near-field optical microscopy (SNOM)[5-7], the confocal microscopy[8,9] and the total internal reflection fluorescence (TIRF) microscopy[10-13]. SMD of diffusing fluorophore molecules were also demonstrated[14, 15]. In 1996, Ha et al. first succeeded the single-molecule FRET (smFRET) measurement of biological molecules by acquiring the fluorescence spectrum using SNOM[16]. Successively, other smFRET experiments, such as the conformational dynamics of proteins in catalytic reactions [17], the ligand-induced conformational change[18] and the protein folding[19], were reported, which indicated the ability of the smFRET measurement to directly observe the dynamics of individual molecules. smFRET measurements of diffusing molecules, which are free from influence by the glass surface, were also demonstrated, suggesting the possibility of detection and analysis of subpopulations [20-23]. In 2000, the smFRET measurement on a live cell was also realized[24]. Then application of smFRET has been extended into the wide range of research on the biomolecular structure and dynamics. A handful of examples will be introduced in the later section.The smFRET measurement enables us to examine individual molecules one by one while the bulk measurement gives only the average. While the bulk measurement cannot distinguish whether only molecules with the middle FRET exist or it consists of the high and the low FRET species, the smFRET measurement tells which is the case. If the FRET distribution is composed of subpopulations, smFRET measurement can resolve them. If the molecular conformation temporally changes, the smFRET measurement traces the dynamics of individual molecules, which is in many cases thermodynamically stochastic and then asynchronous among molecules. At the present time, smFRET is almost the only way to realize such measurement.In the relatively short history of smFRET, which is less than twenty years, superb reviews[25-27] have been already written. They must be helpful for readers to understand the details of smFRET and to build their own instrument. Here this article would be an introductory review of smFRET to summarize basic topics.
2. Theory for smFRET
- When two kinds of fluorophores exist in close proximity to each other and one of them (donor) is in the excited state, the energy can be transferred to the other (acceptor) via nonradiative dipole-dipole interaction without photon emission. That phenomenon is called the FRET. FRET occurs only when the emission spectrum of the donor and the absorption spectrum of the acceptor overlap. The efficiency of energy transfer EFRET was formulated by Förster as[2, 28]
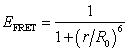 | (1) |
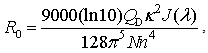 | (2) |
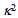 is the orientation factor, which reflects the relative orientation in space of fluorophores and will be mentioned below.
is the orientation factor, which reflects the relative orientation in space of fluorophores and will be mentioned below. 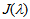 represents the overlap integral, which expresses the degree of spectral overlap between the donor emission and the acceptor absorption. R0 represents the distance, at which the transfer efficiency is 0.5 and is typically 5–10 Å for opt-used dye pairs. The term of 6th power seen in (1) indicates the strong dependence of EFRET on r around R0.EFRET can be calculated as the fraction of the energy transferred to the acceptor in the total energy absorbed by the donor. When fluorescence is detected from a single pair of FRET dyes on two wavelength-separated detector channels, the apparent FRET efficiency Eapp may be approximated with the ratio of detected intensities as
represents the overlap integral, which expresses the degree of spectral overlap between the donor emission and the acceptor absorption. R0 represents the distance, at which the transfer efficiency is 0.5 and is typically 5–10 Å for opt-used dye pairs. The term of 6th power seen in (1) indicates the strong dependence of EFRET on r around R0.EFRET can be calculated as the fraction of the energy transferred to the acceptor in the total energy absorbed by the donor. When fluorescence is detected from a single pair of FRET dyes on two wavelength-separated detector channels, the apparent FRET efficiency Eapp may be approximated with the ratio of detected intensities as 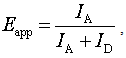 | (3) |
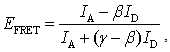 | (4) |
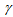 is the coefficient to correct the first factor, the ratio of the detection efficiency between dyes and
is the coefficient to correct the first factor, the ratio of the detection efficiency between dyes and 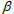 is for compensation of the second factor, the fluorescence leakage[22,29,30].
is for compensation of the second factor, the fluorescence leakage[22,29,30]. 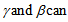 be determined experimentally. If the detected wavelength is restricted by, for example, a band pass filter before the acceptor detector,
be determined experimentally. If the detected wavelength is restricted by, for example, a band pass filter before the acceptor detector, 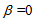 can be assumed at the expense of a portion of photons.
can be assumed at the expense of a portion of photons. 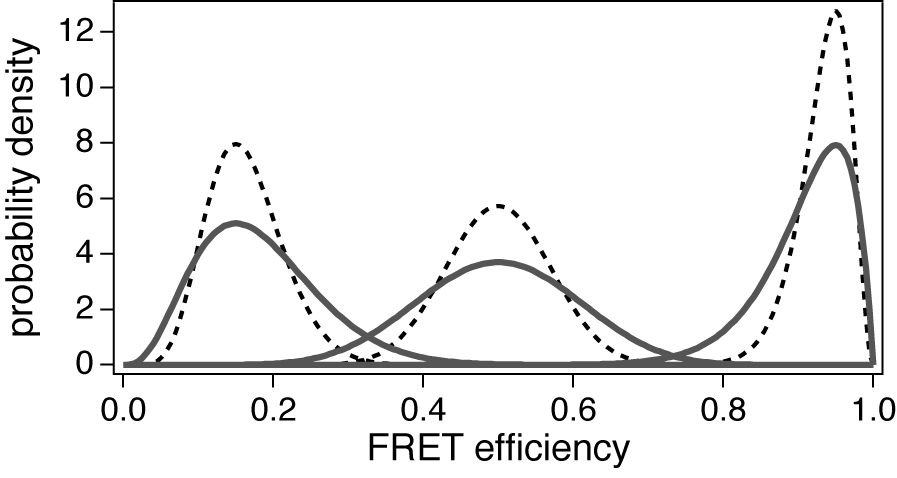 | Figure 1. The theoretical distribution of the FRET efficiency. Solid lines are peaks with I = 20 and E = 0.15, 0.5 and 0.95, respectively. I = 50 for dashed lines |
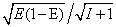 , where E =
, where E = 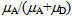 and
and 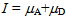 are the mean FRET efficiency and the intensity, respectively, with the average of detected photon counts for the acceptor
are the mean FRET efficiency and the intensity, respectively, with the average of detected photon counts for the acceptor 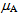 and the donor
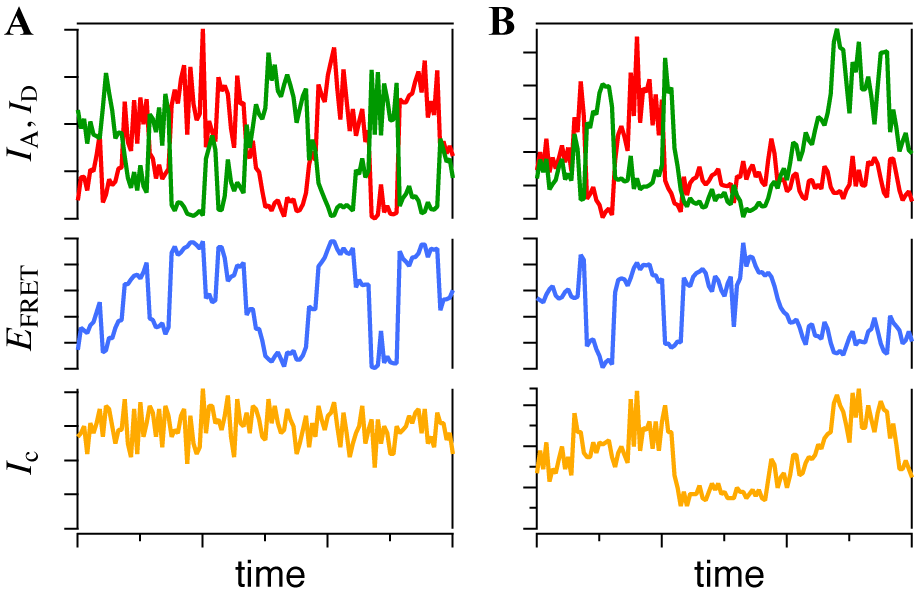
and the donor 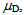 respectively[22]. It suggests that larger I gives a sharper peak and if I is same, the peak becomes sharper near the boundary (E = 0, 1) compared to at the center (E = 0.5). It should be kept in mind that if the compensation of (4) is applied, the Beta distribution describes the distribution before compensation.One of the advantages of smFRET measurement is the ability to trace the time evolution of the FRET changes in realtime like shown in Figures 2. If the FRET efficiency varies with changes in the interdye distance, the acceptor intensity increases while the donor intensity decrease, or vice versa (Figure 2A). However, the fluorescence intensity can be modulated by other reasons than FRET, such as photochemical effects like blinking or photobleach, which are explicitly observed in the single-molecule measurements. When such fluctuations occur, the anti-correlation relationship of the fluorescence intensities is diminished (Figure 2B). In order to distinguish the intensity modulation due to the FRET changes from those instabilities, calculating the cross correlation between two intensity signals was proposed to quantitatively evaluate the anti-correlation relationship[31]. Or more simply, the compensated total intensity Ic can be calculated as[30]
respectively[22]. It suggests that larger I gives a sharper peak and if I is same, the peak becomes sharper near the boundary (E = 0, 1) compared to at the center (E = 0.5). It should be kept in mind that if the compensation of (4) is applied, the Beta distribution describes the distribution before compensation.One of the advantages of smFRET measurement is the ability to trace the time evolution of the FRET changes in realtime like shown in Figures 2. If the FRET efficiency varies with changes in the interdye distance, the acceptor intensity increases while the donor intensity decrease, or vice versa (Figure 2A). However, the fluorescence intensity can be modulated by other reasons than FRET, such as photochemical effects like blinking or photobleach, which are explicitly observed in the single-molecule measurements. When such fluctuations occur, the anti-correlation relationship of the fluorescence intensities is diminished (Figure 2B). In order to distinguish the intensity modulation due to the FRET changes from those instabilities, calculating the cross correlation between two intensity signals was proposed to quantitatively evaluate the anti-correlation relationship[31]. Or more simply, the compensated total intensity Ic can be calculated as[30] 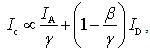 | (5) |
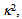 which depends on the relative orientation of two dyes while it is generally difficult to know the actual molecular orientation in real experiments. However, when dyes are labeled via linkers,
which depends on the relative orientation of two dyes while it is generally difficult to know the actual molecular orientation in real experiments. However, when dyes are labeled via linkers, 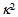 may be approximated to 2/3, which is the theoretical average under the assumption of random orientation. If one can obtain EFRET values for three different FRET states, the relative distance change can be absolutely calculated.The FRET efficiency can be measured from the fluorescence lifetime, too. The fluorescence lifetime is the time lag between the absorption of the excitation light and the fluorescence photon emission on a fluorophore. In order to measure the fluorescence lifetime, which is typically in the order of nanoseconds, a pulsed laser is used for excitation and the decay constant for an exponential distribution of the lag times is calculated from a bunch of photon detection. Then the FRET efficiency can be obtained as
may be approximated to 2/3, which is the theoretical average under the assumption of random orientation. If one can obtain EFRET values for three different FRET states, the relative distance change can be absolutely calculated.The FRET efficiency can be measured from the fluorescence lifetime, too. The fluorescence lifetime is the time lag between the absorption of the excitation light and the fluorescence photon emission on a fluorophore. In order to measure the fluorescence lifetime, which is typically in the order of nanoseconds, a pulsed laser is used for excitation and the decay constant for an exponential distribution of the lag times is calculated from a bunch of photon detection. Then the FRET efficiency can be obtained as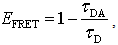 | (6) |
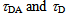 are the fluorescence lifetime of the donor dye with and without FRET, respectively[28].
are the fluorescence lifetime of the donor dye with and without FRET, respectively[28].3. smFRET Methods
- Experimental conditions necessary for the single-molecule experiments are similar to what is required to look at the stars in the sky. First, the background light must be very dark like the night sky. It is essential to achieve a sufficiently high signal-to-noise ratio because each light spot is not very bright. In experiments, care must be taken to keep solvent pure and glass substrates clean. Second, the light spots must be sparsely distributed in order to discriminate each, not like the Milky Way. smFRET samples must be sufficiently diluted so that only a single molecule exists in the spatially resolved detection volume of the microscope. In addition, the optics must be carefully designed to effectively collect fluorescence photons and the detectors must be equipped with the single-molecule sensitivity. Typical detectors used in smFRET experiments are highly sensitive cameras, e.g. EM-CCD and sCMOS, and the single photon counting (SPC) detectors, e.g. a photomultiplier tube (PMT) and an avalanche photodiode (APD). Fluorescence is divided by wavelength, for example, by a dichroic filter, onto two detection channels or its spectrum can be measured by a spectrometer. There are two types of apparatus, which are practically employed for smFRET measurements. They are briefly introduced in the following.
3.1. Solution-phase smFRET
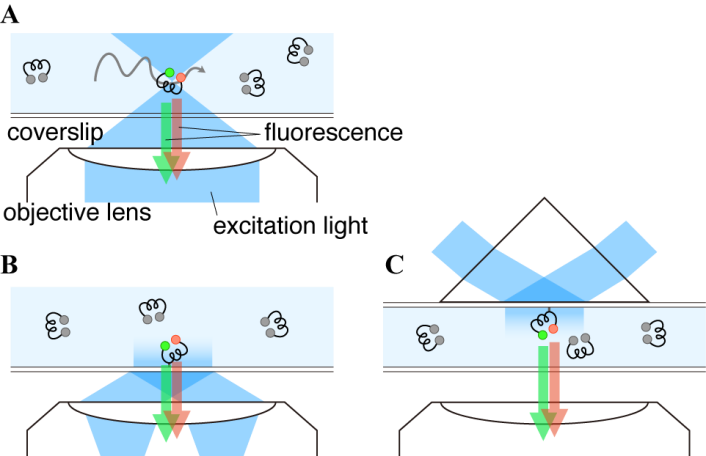
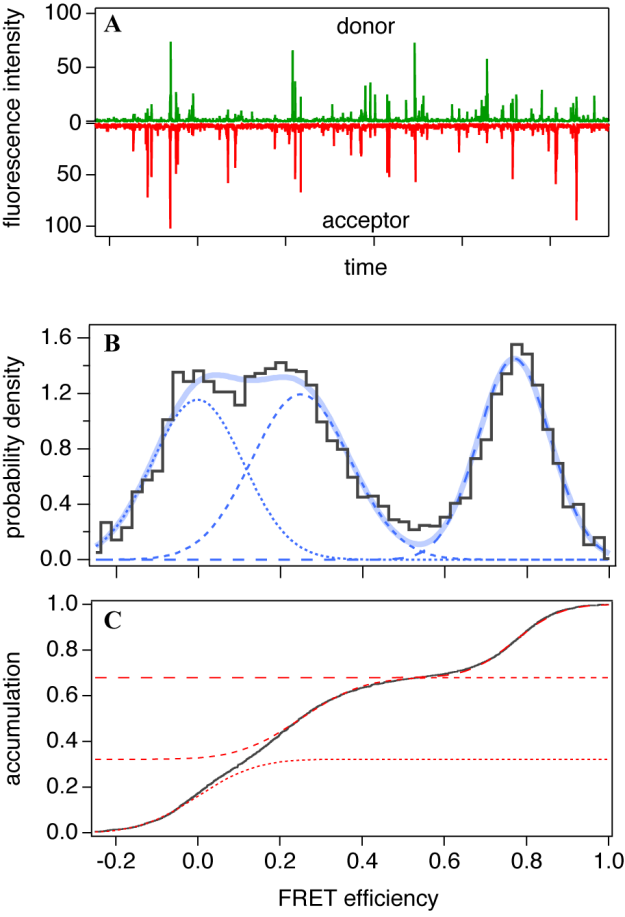
- The dilute solution is a simple way to satisfy experimental requirement of the sparse distribution mentioned above. The focus of a microscope is fixed in a solution (Figure 3A) and then only molecules crossing the focal volume are excited and emit photons. A confocal microscope equipped with SPC detectors is suitable for this type of experiment. Typical time traces of the solution-phase smFRET consist of the low background signal and intermittent fluorescence bursts as shown in Figure 4A. At sufficiently low concentrations, each burst can be regarded as a single molecule. Then the FRET histogram can be constructed by counting photons from each burst (Figure 4B).
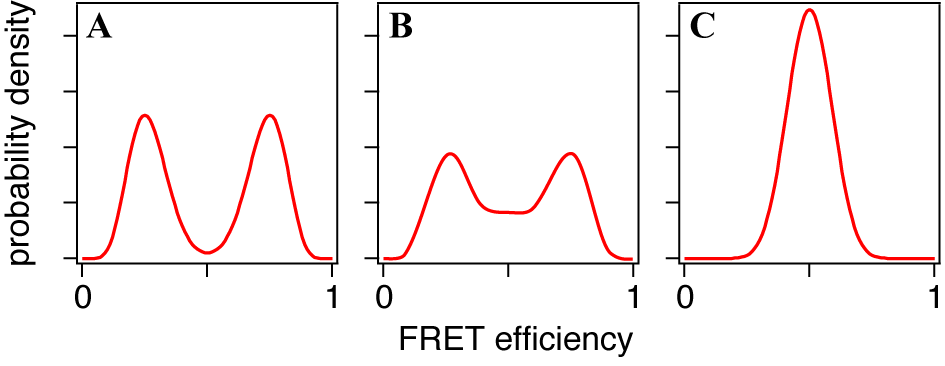 | Figure 5. The typical FRET distributions of the molecules in equilibrium between two conformational states with (A) slow, (B) intermediate and (C) fast transition rates, respectively |
3.2. Surface-immobilized smFRET
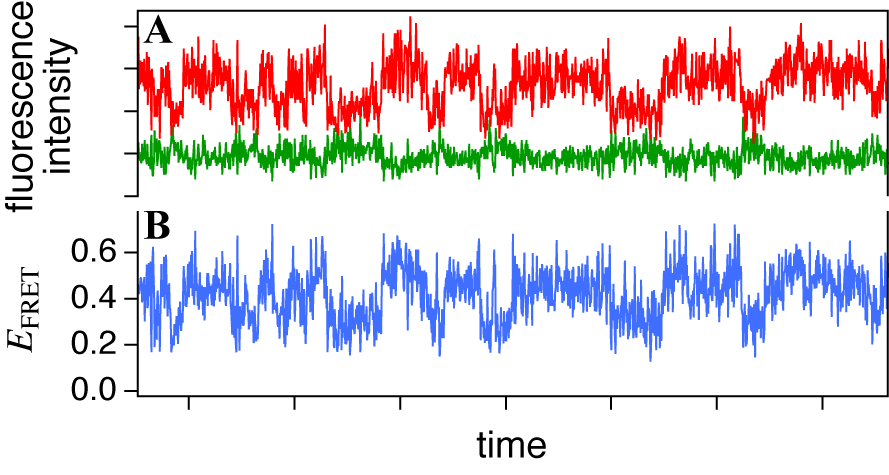
- One of advantages of smFRET measurement is its ability to trace dynamic changes of the molecular structure. In addition to the confocal microscope, the focus of which is adjusted on the substrate surface, the TIRF microscopy is often used (Figure 3B and 3C) to acquire photons from the single molecules immobilized on the surface of the glass substrate. In TIRF microscopy, the evanescent field of the illumination light is generated by reflection on the glass-solution boundary with the larger incident angle than the critical angle. Two types of TIRF configurations, the objective- (Figure 3B) and the prism-type (Figure 3C), are commonly used for that purpose. Since the penetration depth of the evanescent field is typically ~100nm or less, only the surface-bound molecules can be illuminated while suppressing the background signal from solution. Cameras are available with TIRF apparatus while a confocal microscope with SPC detectors achieves the very high signal-to-noise ratio. Examples of the fluorescence time series and the FRET efficiency calculated from them for an immobilized single molecule are shown in Figures 6. The measurement can run until the dye is photobleached. The lifetime of the dye is typically over seconds while it is dependent on various factors, such as solvent composition. The reductant, such as
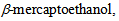 to suppress the fluorescence instability or the oxygen scavenger, such as the combination of glucose, glucose oxidase and catalase, to suppress the photobleach, are often mixed into the solution.
to suppress the fluorescence instability or the oxygen scavenger, such as the combination of glucose, glucose oxidase and catalase, to suppress the photobleach, are often mixed into the solution.4. Data Analysis for smFRET
4.1. Preliminary Filtering
- Sample preparation for smFRET experiments includes many stochastic processes, such as fluorescence labeling and surface immobilization. It is difficult to prepare sample molecules with 100% accuracy. Some molecules may be singly labeled, which may be due to mislabeling or photobleach of either of dyes, and some may be multiply labeled with the same dye. There may be some molecules influenced from the substrate surface because of imperfect blocking. smFRET measurement explicitly detects such unsuccessful molecules as well as fluorescent impurities. Typically, only a fraction of detected molecules gives the meaningful information. Therefore it is desirable to exclude bad molecules before further detailed analysis, such as time-series analysis described below. In the surface- immobilized experiments, since each molecule can be examined with its intensity, FRET efficiency and their temporal changes, one may be able to specify the obviously bad molecules. On the other hand, in solution-phase experiments, it is difficult to discriminate each burst. Distribution analysis described below may help to distinguish them if they form a subpopulation.
4.2. Distribution Analysis
- Once the FRET efficiencies of a bunch of single molecules are measured, their distribution is often summarized as a histogram and devoted to further analyses. When a histogram can be considered to be composed of a few or several components, multiple distribution functions are usually fitted to it. As mentioned in the Section 2, each component can be theoretically represented by a Beta distribution. However, Gaussian functions are practically used to approximate peaks for convenience especially when the signal is not acquired by the SPC detectors because the non-SPC signals are commonly deviated from Poisson statistics. In order to avoid dependence on the binning conditions, a method to fit error functions to the cumulative distribution function without binning was proposed[36] (Figure 4C). Figure 4B shows an example of the smFRET distribution, which is obtained from the mixture of two kinds of dsDNAs. A peak centered at E ~ 0 is typically seen in the experimental smFRET distribution and corresponds to molecules without the acceptor due to mislabeling or photobleach. However, it may possibly include the molecules with the very open structure, the contributions from which are indistinguishable from those from the donor-only molecules in principle. In order to distinguish them, additional information must be acquired, for example, by direct excitation of the acceptor [37].
4.3. Time-series Analysis
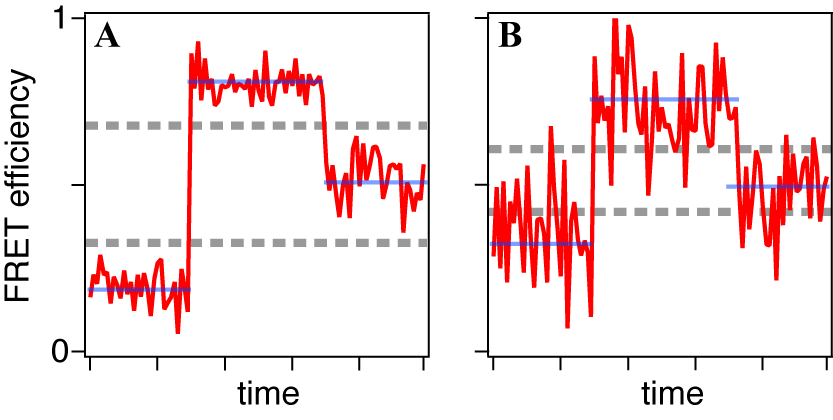
- A smFRET time series obtained from an immobilized molecule gives us the time evolution of conformational dynamics. It often shows stepwise changes as shown in Figures 6 and 7, i.e. the time series consists of successive periods, within each of which the signal level is constant, and instantaneous jumps connecting them. Such trajectory is usually explained by the consecutive transitions among the conformational states. The molecule stays in one of conformational states within a flat period and gives the FRET signal corresponding to that state while the transition dynamics itself completes too fast for time resolution of the smFRET measurement. From that point of view, what we have to do is to divide the trajectory into short periods, each of which corresponds to a stay in a state (blue lines in Figures 7). It is straightforward when the FRET change is large and the fluctuation is small (Figure 7A). States can be discriminated by defining thresholds or can be assigned ‘by eye.’ However, it is not always easy because the smFRET signals usually contain stochastic fluctuations and the FRET change may not be large enough (Figure 7B). In such cases, simple thresholding may cause the ‘artifact’ in detection of the state transitions[38].
5. smFRET Applications to Biomolecules
- smFRET has been extensively applied in the wide range of biological researches. In this section, I would like to introduce only a handful of examples.
6. Conclusions
- The fundamental techniques for the smFRET measurement, including instrumentation, sample preparation and data analysis, have been developed and the practical knowledge has been accumulated in the last two decades. smFRET has been extensively used to investigate biological phenomena, especially dynamic reactions and provided new findings. smFRET has become one of the indispensable tools in life science. On the other hand, it should be kept in mind that, of course, the smFRET measurement is not perfect. It gives structural information only indirectly via the one-dimensional interdye distance. For example, X-ray diffraction crystallography and NMR, which have three-dimensional atomic resolution, are superior in the spatial resolution. Nevertheless, it is an exclusive advantage to possess the single-molecule sensitivity, the nanometer-spatial resolution and the realtimeness, simultaneously. There is certainly the area only smFRET can reach. The smFRET technique will continue to play an important role in biology.
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML