-
Paper Information
- Next Paper
- Previous Paper
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
Science and Technology
p-ISSN: 2163-2669 e-ISSN: 2163-2677
2012; 2(1): 15-20
doi: 10.5923/j.scit.20120201.03
Production of Cellulase and Protease from Microorganisms Isolated from Gut of Archachatina marginata (Giant African Snail)
Oyeleke S. B. 1, Egwim E. C. 2, Oyewole O. A. 1, John E. E. 1
1Department of Microbiology, Federal University of Technology, Minna, Niger State
2Department of Biochemistry, Federal University of Technology, Minna, Niger State
Correspondence to: Oyeleke S. B. , Department of Microbiology, Federal University of Technology, Minna, Niger State.
| Email: |  |
Copyright © 2012 Scientific & Academic Publishing. All Rights Reserved.
Cellulase and protease producing microorganisms were isolated from the gut of Archachatina marginata (Giant African snail). The microorganisms isolated were Bacillus subtilis, Staphylococcus aureus, Streptococcus casseliflavus, Streptococcus faecalis, Aspergillus flavus, Rhizopus sp, Fusarium sp. and Aspergillus niger. When the isolates were tested on skimmed milk agar (1%) and Carboxymethyl Cellulase, B. subtilis and A. niger were the best producing microorganisms for both cellulase and protease enzyme. For cellulase production, B. subtilis revealed its highest biomass yield after 24 hours of incubation with activity of 0.62mg/ml/secˉ4 at 80℃ and pH of 5. Aspergillus niger showed its highest biomass yield after 5 days with an activity of 0.74mg/ml/secˉ4 and an optimum temperature of 60℃ and the optimum pH was 5. For protease production, B. subtilis was grown for 30 hours; its highest biomass yield was after 18 hours with an enzyme activity of 2.95µg/ml/secˉ4 and an optimum temperature of 60℃, while A. niger was grown for 7 days and showed highest yield after 6 days with enzymatic activity of 3.14µg/ml/secˉ4, and an optimum temperature of 50℃. The optimum pH was at 7 and 8 for B. subtilis and A. niger respectively. The results from this study suggest that B. subtilis and A. niger can be harnessed for the production of cellulase and protease.
Keywords: Archachatina marginata, Cellulase, Protease, Microorganisms, Enzymatic Activity
Cite this paper: Oyeleke S. B. , Egwim E. C. , Oyewole O. A. , John E. E. , "Production of Cellulase and Protease from Microorganisms Isolated from Gut of Archachatina marginata (Giant African Snail)", Science and Technology, Vol. 2 No. 1, 2012, pp. 15-20. doi: 10.5923/j.scit.20120201.03.
1. Introduction
- Cellulose is a fibrous, insoluble, crystalline polysaccharide. It is a major polysaccharide constituent of plant cell walls, composed of repeating D-glucose units linked by α-1,4-glucosidic bonds (Jagtap and Rao, 2005) and being the most abundant carbohydrate polymer on earth (Guo et al., 2008). Cellulose is used as a food source by a wide variety of organisms including fungi, bacteria, plants and protists, as well as a wide range of invertebrate animals, such as insects, crustaceans, annelids, mollusks and nematodes (Watanabe and Tokuda, 2001; Davison and Blaxter, 2005). These organisms possess cellulases and the complete enzymatic system of them include three different types, thatis, exo-α-1,4-glucanases (EC 3.2.1.91), endo-α-1,4-glucanases (EC 3.2.1.4), and α-1,4-glucosidase (EC 3.2.1.21) (Wilson and Irwin, 1999). Protease (serine protease) {EC 3.4.21}, cystein (thiol) protease (EC 3.4.22), aspartate proteases (EC 3.4.23), and metallo-protease (EC 3.4.24) constitute one of the most important groups of industrial enzymes, accountingfor about 60% of the total enzyme market (Wellingta et al., 2003). Protease is an enzyme that breaks the peptide bonds of proteins (Mitchell et al., 2007). Protease breaks down peptide bonds to produce amino acids and other smaller peptides. It can be isolated from a variety of sources such as plants, animals and microbiota (fungi and bacteria). Its application is very broad and has been used in many fields for years, and is mainly used in food and detergent industries (Yandri et al., 2008). They find application in a number of biotechnological processes, viz. in food processing and pharmaceuticals, leather industry, detergent industry, etc. (Nascimento and Martins, 2004; Beg and Gupta, 2003).Two third (2/3) of the industrial produced proteases are from microbial sources (Ellaiah and Adinarayana, 2002). A variety of microorganisms such as bacteria, fungi, yeast and Actinomycetes are known to produce these enzymes (Madan et al., 2002; Devi et al., 2008). Molds of the genera Aspergillus, Penicillium and Rhizopus are especially useful for producing proteases, as several species of these genera are generally regarded as safe (Devi et al., 2008). Among the various proteases, bacterial protease is the most significant, compared with animal and fungal protease (Wellingta et al., 2004), and among bacteria species. Snails contain a lot of organisms in its gut, and a variety of enzymes can be extracted and produced from it. Snails are cheap and can be easily obtained, especially during the rainy season. The dependence of snails on microbial activity within their gut would explain their extraordinary efficiency in plant fibre digestion (60-80%) (Charrier et al., 2006). In snails, the enzyme producing symbiotic protozoan present in its intestine (Charrier et al., 2006) hydrolyze cellulose to glucose in synergistic collaborative relationship with indigenously produced enzymes (Chevalier et al., 2003). Cellulase and protease can be isolated, extracted and produced locally, thereby saving the cost of importation into the country. These enzymes are especially useful in biofuel industry, fruit juice clarity in fruit juice industry, paper and pulp industry, and biological detergents. Cellulase and protease are important commercial enzymes and the best way of producing these enzymes is via microbial fermentation which is very economical and convenient. This is because of their usefulness in pharmaceutical, paper, detergent industries and many others. The study is aimed at isolating microorganism present in gut of snails (Archachatina marginata) for their potential to produce cellulase and protease and to determine the optimum temperature and pH for the enzyme activities.
2. Materials and Methods
- Collection of SamplesThe snails (5 in number) were obtained from Kure Ultra Modern market in Minna, Niger State, Nigeria. They were placed in a clean, covered bowl, containing sand, some vegetables and water and kept safe in the microbiology laboratory of Federal university of Technology, Minna, Niger State, Nigeria prior to its use. MediaMedia used were Sabouraud Dextrose Agar for isolation of fungi and Nutrient agar for isolation of bacteria. The media were prepared according to manufacturer’s instruction and sterilized in autoclave at 121℃ for 15 minutes.Isolation of the Microorganisms from the gut of Archachatina marginataThe snails were washed with clean water and the outer shell cleaned thoroughly with ethanol for surface sterilization. The shells were aseptically broken to remove its flesh and then dissected to reveal its gut. A portion of the gut was streaked onto nutrient agar plates, using inoculating loop and incubated for 18-24hours at 37℃ for bacterial growth and streaked in Sabouraud dextrose agar (SDA) plates and incubated for 25℃ for 3-5days for fungal growth. Individual colonies observed were sub-cultured on Nutrient agar and Sabouraud dextrose agar plates for another 18-24 hours and 3-5 days respectively and finally grown on agar slants to preserve the pure cultures.Identification of MicroorganismsThe bacterial isolates were characterized based on colonial morphology, cultural characteristics and biochemical tests as described by Oyeleke and Manga (2008). The isolates were identified by comparing their characteristics with those of known taxa using the Bergey’s manual of determinative bacteriology (Holts et al., 1994). A small portion of the mycelia growth was carefully picked with the aid of a pair of sterile inoculating needles and placed in a drop of lactophenol cotton blue on a microscope slide and covered with a cover slip. The slide was examined under the microscope, first with (x10) and then with (x40) objective lens for morphological examination as described by Oyeleke and Manga, (2008). The isolates were identified by comparing their characteristics with those of known taxa using the schemes of Domsch and Gams (1970).Screening for Protease EnzymeOne gram (1g) of powdered milk was included into Nutrient Agar and Sabouraud dextrose agar respectively and the bacteria and fungi isolates were inoculated unto the plates, for 24 hours at 37℃, and 5 days at room temperature (28±2℃) respectively. The bacteria plates were observed for a zone of clearance after 24hours and 5days for fungi plates.Screening for Cellulase ActivityA loopful of grown culture of isolated colonies was inoculated on Nutrient broth containing 1% Carboxymethyl Cellulose (CMC) for bacteria in conical flasks, and Potato dextrose broth also containing 1% Carboxymethyl Cellulose (CMC) for fungi. They were incubated at 37℃ for 24hours for bacteria isolates and for 5days at room temperature (28±2℃) for fungi isolates and allowed to grow. After incubation, the cellulolity activity was measured using spectrophotometer by dinitrosalicylic acid (DNSA) method (Bertrand et al., 2004). This was done by centrifuging the culture broth at 8, 000 revolutions per minute (rpm) for five (5) and fifteen (15) minutes for bacteria and fungi respectively. It was then filtered using Whatman’s filter paper No. 1 and then one ml (1 ml) of the culture filtrate, one ml (1 ml) 1% CMC in 0.05M citrate buffer, pH 4.8 incubated for 30minutes at 37℃ then reacting with DNS reagent (to stop the growth reaction) and boiled for 5 minutes. It was then read with the spectrophotometer at 540 nM.Cellulase Enzyme Production and AssayThe bacteria species was grown for production of cellulase in a minimum salt medium, (500ml) containing 0.05g MgSO4 , 0.5g (NH4)2SO4, 1g KH2PO4, 3.5g K2HPO4, 0.25g Sodium citrate, supplemented with carboxymethyl cellulose (CMC) as carbon source in distilled water (it is sterilized at 121℃ for 15 minutes, allowed to cool before use) . The cultures were grown at 37℃ for 30 hours. Culture broth was sampled at a six (6) hour interval during growth to enzyme activity in relation to biomass yield by measuring at an absorbance of 540 nM using, spectrophotometer. Cellulase activity was assayed by the determination of reducing sugar released from carboxymethyl cellulose (CMC). 0.5 ml of culture supernatant fluid was incubated with 0.5 ml 1% CMC in 0.05M citrate buffer, pH 4.8 at 40℃ for 30mins. The reducing sugar product was assayed by the dinitrosalicylic acid (DNSA) method (Bertrand et al., 2004), using glucose as the sugar standard. The fungi species were placed in a minimal salt medium: FeSO4 0.001g; NaNO3, 1.5g; Na2HPO4, 0.5g; MgSO4, 0.05; and KCl; 0.25g; Carboxymethyl cellulose (CMC); 5g in 500ml capacity distilled water (it was sterilized at 121℃ for 15 minutes and allowed to cool before use). The culture was grown for 7days at room temperature (28±2℃). The culture broth was sampled every 24 hours during growth to determine cell density/biomass growth by measuring at an absorbance of 540 nM, using spectrophotometer. Cellulase assay was done according to the method of Mandels (2005). Half milliliter of 1% CMC in 0.1M citrate buffer (pH 4.8) was placed in a test tube and 0.5 ml of culture filtrate was added. The reaction mixture was incubated at 50℃ for 30 min and the reaction terminated by adding 1.5 ml 3,5- dinitrosalisylic acid (DNSA) reagent. The tubes were heated at 100℃ in a boiling water bath for 5 minutes and then cooled at room temperature (28±2℃). The absorbance was read at 540 nM. Enzyme activity is expressed as mg/ml glucose released per min-1ml of culture filtrate as enzyme solution.Culture filtrate was obtained by filtration through What- man No. 1 filter paper and the culture filtrate served as the enzyme solution (Singh, 2003; El-Naghy et al., 1991).Protease Enzyme production and AssayThe bacteria isolate was grown in culture medium containing 2.5g glucose, 3.75g peptone, 2.5g MgSO4, 2.5g KH2PO4, 0.05g FeSO4, 3g Casein in 500ml distilled water in a conical flask (it is sterilized at 121℃ for 15 minutes, allowed to cool before use) and incubated for 24hours. The fungal isolates were grown in KH2PO4, 0.5g; MgSO4·7H2O, 2.5g; KCl, 2.5g; FeSO4·7H2O, 0.05; glucose, 2.5g; peptone, 3.75g; casein, 5g in 500ml distilled water in a conical flask (it is sterilized at 121℃ for 15 minutes, allowed to cool before use) for protease production. The culture broth for bacteria is sampled every six (6) hours, during growth and that of fungi every 24 hours during growth to determine the enzyme activity in relation to biomass growth for protease enzyme. The protease activity was assayed by the method of Lovrien et al., (1985). Three ml (3 ml) of reaction mixture containing 0.5% casein in 2.95 ml of 0.1 M Tris-HCl buffer, pH 8.0 and 0.1 ml of each enzyme was incubated at 40℃. After 30 min, the reaction was stopped by adding 3 ml of cold 10% trichloroacetic acid. After 1 h, each of the culture filtrate was centrifuged at 8,000 rpm for 5 min to remove the precipitate and absorbance of the supernatants was read with spectrophotometer at 540 nm. The amount of amino acids released was calculated from a standard curve plotted against a range of known concentrations of tyrosine. One unit of enzyme (µ/ml/min) was defined as the amount of enzyme that liberated 1.5 g tyrosine per ml per minute under assay conditions (Bertrand et al., 2004).Effect of pH on the Production of CellulaseThe optimum pH was determined by incubating 1 ml appropriately crude enzyme, 1ml 1% CMC in 1 ml citrate buffer, pH 4.8 (Sami et al., 2008) different pH (4 – 9) for 30mins at 50℃. Reducing sugars thus released were estimated by the dinitrosalicyclic acid reagent method (Bertrand et al., 2004).Effect of Temperature on the Production of CellulaseThe optimum temperature of CMCase was determined by incubating 1 ml appropriately diluted enzyme with 1 ml 1% CMC in citrate buffer pH 4.8 (Sami et al., 2008) at different temperature range (30 – 80℃) for 30mins. Reducing sugars were estimated by the dinitrosalicyclic acid reagent method (Bertrand et al., 2004).Effect of pH on the Production of ProteaseThe effect of pH on protease production was carried out by growing both fungi and bacteria species at different pH range (4-9). Protease activity was measured using spectrophotometer. Effect of Temperature on the Production of ProteaseThe optimum temperature of protease produced was studied by incubating 1 ml crude enzyme, 1 ml 1% CMC in citrate buffer, pH 5 growing both fungi and bacteria species at various temperature range (40-90℃). Protease activity was measured using spectrophotometer.
3. Results
- Screening of Bacterial Isolates for the Production of CellulaseFive bacteria were isolated from the gut of the snail. Of the four (4) bacterial isolated and screened for cellulase enzyme production, B. subtilis showed the highest cellulase activity (2.2 mg/ml/sec-4) while S. aureus showed the least (Table 1).
|
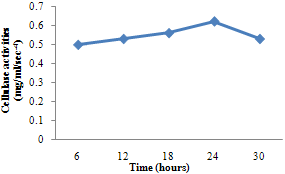 | Figure 1. Cellulase activity of B. subtilis |
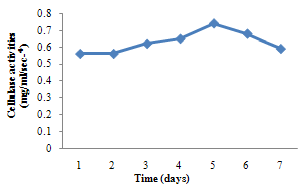 | Figure 2. Cellulase activity of A. niger |
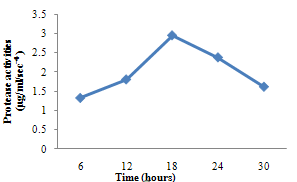 | Figure 3. Protease activity of B. subtilis |
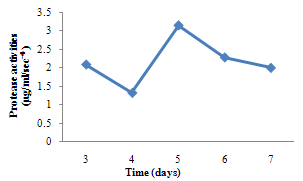 | Figure 4. Protease activity of A. niger |
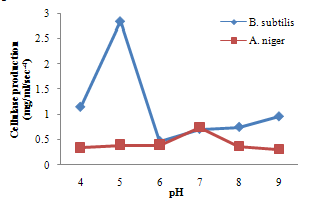 | Figure 5. Effect of pH on cellulase production |
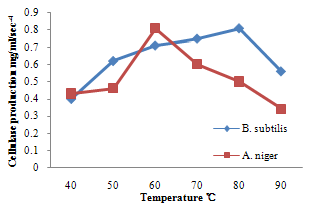 | Figure 6. Effect of temperature on cellulase production |
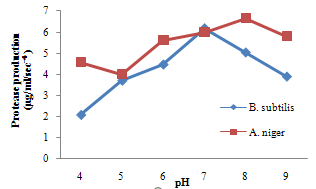 | Figure 7. Effect of pH on Protease production |
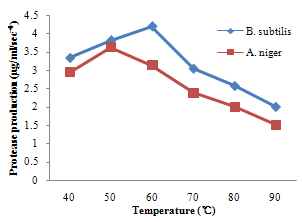 | Figure 8. Effect of temperature on protease production |
4. Discussion
- For cellulase enzyme, B. subtilis was cultivated in mineral salt medium for 30 hours, at 37℃. After 24 hours, it showed its highest enzymatic activity to produce cellulase at 0.62mg/ml/sec-4. These are in correlation to the findings of Mohamed et al. (2010) who recorded maximum cellulase productivity after 24 hours for B. subtilis KO strain. Aspergillus niger was also cultivated in mineral salt medium for 7 days at room temperature, and it showed its highest enzymatic activity after 5 days with an activity of 0.74×104 mg/ml/sec. These were slightly above findings by Umbrin et al. (2011) that recorded maximum cellulase productivity after 4 days in the solid state fermentation of A.niger by Vigna mungo. Different experiments, such as temperature and pH were carried out to optimize the culture conditions for the enzyme produced. B. subtilis had an optimum pH of 5, other Bacillus species, according to previous studies, by Ogura et al. (2006) had an optimum pH range of 5-6.5. The optimum temperature was at 80℃, however, this is in contrast with the findings of Umbrin et al. (2011) who recorded an optimum temperature of 45℃ for B. subtilis KO strain and B. Amyloliquefaciens. Ahmeh et al. (2004) and Kahan et al. (2006) recorded an optimum temperature of 40℃ for Bacillus strains DLG but Howard et al. (2003) recorded a temperature value of 65℃ as optimum for cellulase activities by Bacillus sp. The optimum pH for A. niger was 7 which is similar to the findings of Ogura et al. (2006), for other Aspergillus species with an optimum pH of 6-7. The optimum temperature was 60℃. This value is higher than that reported for Streptomyces sp. (Alam et al., 2004) with optimum temperature of 50℃, but lower than that reported for endoglucanase from Aspergillus niger (Howard et al, 2003) with optimum temperature of 70℃.For protease enzyme, B. subtilis exhibited its highest enzymatic activity after 18 hours with an activity of 2.95×104 µg/ml/sec after cultivating in mineral salt medium for 30 hours. Although, this is different from findings by Wellingta et al.(2004) that had maximum enzymatic after 9 hours for Bacillus sp. A. niger had its highest enzymatic activity in 6 days, with an activity of 3.14×104 µg/ml/sec after cultivation in mineral salt medium for 7 days. B. subtilis had an optimum pH of 7, this is quite similar to the findings of Wellingta et al. (2004) that reported an optimum pH of 8 and optimum temperature of 60℃. A. niger recorded an optimum pH of 8, similar to findings by Oyeleke et al. (2010), and an optimum temperature of 50℃. Thus, B. subtilis and A. niger isolated from gut of A. marginata can potentially produce cellulase and protease and can be exploited for the commercial production of the enzymes in industries.
References
| [1] | Alam, M.Z., Manchur, M.A and Anwar, M.N. (2004). Isolation, purification and characterizationof cellulolytic enzymes produced by the isolate Streptomyces omiyaenisis. Pakistan Journal of Biological Sciences 7(10) 1647-1653 |
| [2] | Ahmeh, T., Aslam, Z., and Rasool, S. (2004). Reducing fibre content of sunflower oil meal through treatment of enzyme produced from Archnoitus sp. Animal Science Journal 75:231Beg |
| [3] | Beg, K.B., and Gupta, R. (2003). Purification and characterization of an oxidation-stable, thiol-dependent serine alkaline protease from Bacillus mojavensis. Enzyme and Microbiology Technology, 32: 294-304 |
| [4] | Bertrand, T.F., Frederic T. and Robert N. (2004). Production and Partial Characterization of a thermostable amylase from Ascomycetes yeast strain isolated from starchy sail. McGraw Hill Inc, New York. pp. 53-55 |
| [5] | Charrier, M.Y., Gerard F., Gaillard-Martinie, B., Kader, A., and Gerard, A. (2006). Isolation and characterization of cultivable fermentative bacteria from the intestine of two edible snails, Helixpomatia and Cornuaspersum (Gastropoda: pulmonata). Biological Research 0716-9760 |
| [6] | Chevalier, L., Le Coz-bouhnik, M., and Charrier, M. (2003) Influence of inorganic compounds on food selection by the brown garden snail Cornu aspersus (Miiller) (Gastropoda pulmonata). Malacologia 45: 125-132 |
| [7] | Davison, A., and Blaxter, M. (2005). Ancient origin of glycosyl hydrolase family 9 cellulase genes. Molecular Biology Evolution 22(5): 1273-1284 |
| [8] | Devi, M.K., Banu, A.R., Gnanaprabhal, G.R., Pradeep B.V. and Palaniswamy M. (2008). Purification, characterization of alkaline protease enzyme from native isolate Aspergillus niger and its compatibility with commercial detergents. Indian Journal of Science Technology, 1: 7 |
| [9] | Domsch, K.H. and Gams, W. (1970). Fungi in Agricultural Soils (First Edition). Longman Group Limited, London, pp. 20 - 152 |
| [10] | Ellaiah, C.A., and Adinarayan, M.L. (2002). Production and properties of an extracellular protease from Bacillus sp. Journal of Microbiology, 35: 91-96 |
| [11] | El-Naghy, M.A., El-Katatny, M.S. and Attia, A.A. (1991). Degradation of cellulosic materials by Sporotrichum thermophile culture filtrate for sugar production. International Biodeterioration, vol. 27, no. 1, p. 75-79 |
| [12] | Guo, R., Ding, M., Zhang, S.L., Xu, G.J., and Zhao, F.K. (2008). Molecular cloning and characterization of two novel cellulase genes from the mollusk Ampullaria crossean. Journal of Compressed Physiology [B], 178(2): 209-215 |
| [13] | Holt, J.G. Krieg, N.R., Sneath, P.H.A., Staley, J.T. and Williams, S.T. (1994). Bergy’s Manual of Determinative Bacteriology. Williams and Wilkins Publishers, Maryland |
| [14] | Howard, R.L., Masoko, P. and Abotsi, E. (2003). Enzyme activity of a Phanerochaete chrysosporium cellobiohydrolase (CBHI.1) expressed as a heterologous protein from Escherichia coli African Journal of Biotechnology Vol. 2 (9), pp. 296-300 |
| [15] | Jagtap, S., and Rao, M. (2005). Purification and properties of a low molecular weight 1,4-beta-d-glucan glucohydrolase having one active site for carboxymethyl cellulose and xylan from an alkalothermophilic Thermomonospora sp. Biochemist and Biophysics Resource Communication 329(1):111-116 |
| [16] | Kahan, F.A.A., and Husaini, A.S.S. (2006). Enhancing α-Amylase and Cellulase in vivo enzyme expressions on sago pith residue, using Bacillus amyloliquefaciens, Journal of Biotechnology, 5: 391-403 |
| [17] | Lovrien RE, Gusek T, Hart B (1985). Cellulase and protease specific activities of commercially available cellulase preparations. J. Appl. Biochem., 7: 258-272 |
| [18] | Madan, M., Dhillon, S, and Singh, R. (2002). Production of alkaline protease by a UV mutant of Bacillus polymyxa. Indian Journal of Microbiology 42: 155-159 |
| [19] | Mandels, M. (2005). Applications of cellulases. Biochemical Society Transactions. (13) 414-416 |
| [20] | Mitchell, R.S., Kumar, V., Abbas, A.K., and Fausto, N. (2007). Robbins Basic Pathology. 8th edition, 122 |
| [21] | Mohamed, S.A., Magadi, A.M., Francis, F.H, and Moustafa, A.N. (2010). Production of cellulase in low-cost medium by Bacillus subtilis KO Strain. World applied Science Journal, 8(1):35-42 |
| [22] | Nascimento, W.C.A., Martins, M.L.L., (2004). Production and properties of an extracellular protease from thermophilic Bacillus spp. Brazilian Journal of Microbiology 35: 1-2 |
| [23] | Ogura, J., Toyoda, A., Kurosawa, T., Chong, A. L., Chohnan, S, and Masaki, T. (2006). Purification, characterization, and gene analysis of cellulose (Cel8A) from Lysobacter sp. IB-9374. Bioscience, Biotechnology, and Biochemistry, (70) 2420-2428 |
| [24] | Oyeleke S. B. and Manga B. S. (2008). Essentials of Laboratory Practicals in Microbiology First Edition, Tobest Publisher, Minna, Niger State, Nigeria pp. 15-34 |
| [25] | Oyeleke, S.B., Auta, S.H, and E. C. Egwim, E.C. (2010). Production and characterization of amylase produced by Bacillus megaterium isolated from a local yam peel dumpsite in Minna, Niger State. Journal of Microbiology and Antimicrobials, 2(7):88-92 |
| [26] | Sami, A.J., Yasmeen N., Shakoori, A.R. (2008) Cellulolytic activity of microbial flora of agricultural insects. Pakistan Journal of Zoology; 4(1):60 – 3 |
| [27] | Singh, C. J. (2003). Optimization of an extracellular protease of Chrysosporium keratinophilum and its potential in bioremediation of keratinic wastes. Mycopathologia, 156: 151- 156 |
| [28] | Umbrin, I., Abdul, M., Khalid, H., Khalid, N., Skakil, A, and Muhammad N. (2011). Solid state fermentation of Vigna mungo for cellulase production by Aspergillus niger. World Applied Sciences Journal, 12(8):1172-1178 |
| [29] | Watanabe, H., and Tokuda, G. (2001). Animal cellulases. Cellular Molecular Life Sciences 58(9):1167-1178 |
| [30] | Wellingta, C. A. D. N., and Meire, L.L.M. (2004) Production and properties of an extracellular protease from thermophilic bacillus sp. Brazilian J. Microbiol. (35): 91-96 |
| [31] | Wilson, D.B., and Irwin, D.C. (1999). Genetics and properties of cellulases. Advances in Biochemical Engineering/ Biotechnology: Recent Progress in Bioconversion, 65: 1-21 |
| [32] | Yandri, A.S., Suhartati, T., Herasari, D. and Hadi, S. (2008). The chemical modification of protease enzyme isolated from local bacteria isolate Bacillus subtilis 1TBCCB148 with cynauric chloride – poly –ethyleneglycol. European Journal of Science Resources 23(1):177-186 |
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-Text HTML
Full-Text HTML