-
Paper Information
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
Research In Cancer and Tumor
2015; 4(1): 15-23
doi:10.5923/j.rct.20150401.03
Discordant Reports of miRNA Expression in Cervical Cancer: An Upshot of Overlapping Factors
Okoye Jude Ogechukwu
Department of Medical Laboratory Science, Faculty of Health Science, Madonna University, Elele Campus, Nigeria
Correspondence to: Okoye Jude Ogechukwu, Department of Medical Laboratory Science, Faculty of Health Science, Madonna University, Elele Campus, Nigeria.
| Email: |  |
Copyright © 2015 Scientific & Academic Publishing. All Rights Reserved.
Background: miRNAs are a class of small non-coding RNAs that are approximately 21-24 nucleotides in length and regulate gene expression post-transcriptionally by way of translational repression or transcript cleavage. They are undoubtedly dysregulated in several neoplastic events but the discordant reports trailing some of them have limited their application in diagnostic pathology.The aim of this review was to evaluate reports of miRNA dysregulation in cervical cancer in a bid to finding explanation for the discordance andtoidentify the best diagnostic and prognostic biomarker for cervical cancer.Method: Articles on miRNA expression in cervical cancer were downloaded from journal websites, Google scholar, Pubmed and Researchgate. The MiRNAs were classified using two criteria; 1) Frequency of reports using a scale of: 6-5 for Class I, 4-3 for Class II, 2 for Class III and 1 report for Class IV; 2) status of controversy, which included: Controversial (C) and Uncontroversial (UC). The controversial group was subdivided into Most Controversial (MC; >25%) and Less Controversial (LC; ≤25%). Result: Sixty-six (66) miRNAs were found to be dysregulated in cervical cancers, out of which 9 (13.6%) miRNAs were controversial, 20 (30.3%) miRNAs were uncontroversial and 37 (56.1%) miRNAs are yet to be validated by other reports. Considering frequency of reports, 5 miRNAs (7.7%), 13 miRNAs (20%), 10 miRNAs (15.4%) and 37 (56.9%) were grouped into Class I, II, III and IV, respectively. In all, only 8 (12%) miRNAs (miR-21, miR-18a, miR-20a, miR-29a, miR-25, miR-183, miR-196a, miR-218) were found to be most reported and without controversy. Conclusions: the expression of miRNAs is affected by overlapping factors such as cancer stage, type of sample analyzed, technique employed, type of viral agent and oncoprotein present. In is important that this factors be considered prior to sample analysis. Since miR-21 is more frequently reported without any discordance when compared with other miRNAs, it should be adopted as the best biomarker in monitoring and management of cervical cancer patients.
Keywords: Cervical cancer, miRNA discordance, miRNA Reports, Viral agent, Oncoprotein
Cite this paper: Okoye Jude Ogechukwu, Discordant Reports of miRNA Expression in Cervical Cancer: An Upshot of Overlapping Factors, Research In Cancer and Tumor, Vol. 4 No. 1, 2015, pp. 15-23. doi: 10.5923/j.rct.20150401.03.
Article Outline
1. Introduction
- Cervical cancer is the third most common cancer in women around the world, with an estimated 530,000 new cases in 2008 [1, 2]. On world-wide basis, cervical cancer ranges from 6 to 9.8% of all cancer in women [1-3]. High-risk regions are Eastern and Western Africa; with greater than 30 cases per 100,000 inhabitants [2]. About 80% of the cases are found in developing countries, which have only 5% of global cancer resources [4-6]. MicroRNAs (miRNAs) are a group of short non-coding RNAs that ranges from 21-24 nucleotides in length and regulate gene expression post-transcriptionally by translational repression or transcript cleavage [7-9]. Many studies have shown that miRNAs are related to cell processes [7, 9], and the dysregulation of these events can lead to the occurrence of a number of diseases, such as prostate cancer [10], and cervical cancer [11] amongst others. MicroRNAs (miRNA) act by inhibiting messenger RNA (mRNA) in several biological processes, including proliferation, differentiation, development, metabolism and cell death. The functional disorders of miRNA are caused by gene mutation, epigenetic variation or genetic polymorphism [12]. However, some controversies have been observed in the reports of some researchers on miRNAs expression in cancers, particularly cervical cancer. Some reports have it that some miRNAs are over expressed in cervical cancer while others suggest otherwise. The difference in reports could be adduced to several factors. The aim of this review was to evaluate reports of miRNA dysregulation in cervical cancer in a bid to finding explanation for the discordance in reports and to identify the best diagnostic and prognostic biomarker for cervical cancer.
2. Methods
2.1. Data Collection and Collation
- This study was carried out on the findings on research papers involving 66 miRNAs (miR-7, miR-10a, miR-17, miR-18b, miR-24, miR-26a, miR-27b, miR-29c, miR-92, miR-106, miR-106b, miR-125, miR-126, miR-132, miR-133a, miR-133b, miR-135b, miR-146, miR-148a, miR-149, miR-181c, miR-193b, miR-200c, miR-205, miR-210, miR-224, miR-301b, miR-375, miR-376a, miR-379, miR-429, miR-449a, miR-500, miR-503, miR-513, miR-522, miR-1293, miR-9, miR-15b, miR-16, miR-10b, miR-27a, miR-29a, miR-31, miR-100, miR-141, miR-155, miR-424, miR-18a, miR20a, miR-20b, miR-25, miR-34a, miR-125b, miR-127, miR-182, miR-183, miR-21, miR-106a, miR-143, miR-145). The research articles were collected from several journal websites and search engines such as Google search, Researchgate and Pubmed central.
2.2. Classification of miRNAs
- MiRNAs were classified using two criteria; 1) Frequency of reports using a rate of: 6-5 for Most Reported (Class I), 4-3 for More Reported (Class II), 2 for Less Reported (Class III) and 1 report for unvalidated (Class IV; 2) status of controversy which include: Controversial (C), Uncontroversial (UC). The controversial group was further subdivided into Most Controversial (MC) and Less Controversial (LC). The most controversial miRNAs were sub-classified using the number of upregulated reports over the entire reports; ½, ⅓ and 3/6 (>25% discordance) while the less controversial miRNAs were classified the same but with minimal degree of controversy ¼, 1/5 and 1/6 (≤25% discordance).
2.3. Interactions between Frequency and Controversy of miRNA Reports
- A set was use to show the relationship between frequency and status of controversy (controversial and uncontroversial) for the identified 66 miRNAs.
3. Result
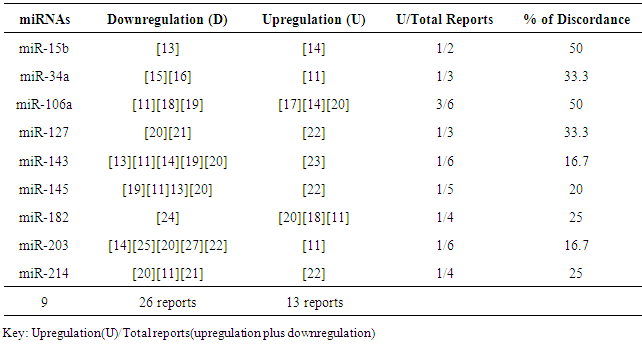
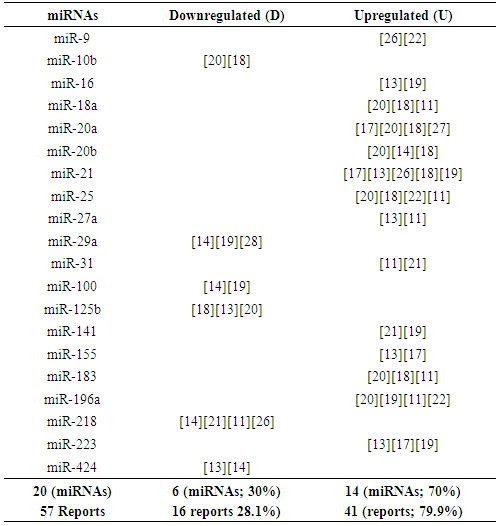
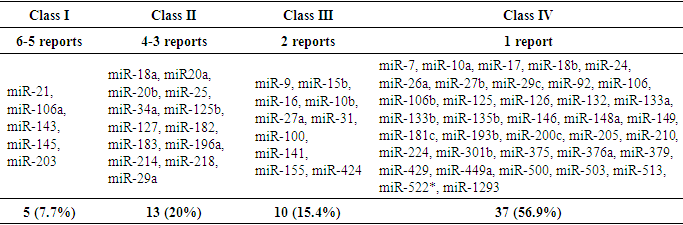
- Table 1 shows the 9 controversial miRNAs as found in some reports. Twenty-six (26) studies reported downregulation (under expression) of the miRNAs while 13 studies reported upregulation (over expression) of the miRNAs. This means that the reports of downregulation (tumor suppressor miRNAs) are twice the reports of upregulation (oncogenic miRNAs).Table 2 shows identified uncontroversial 20 miRNA expressions in literature. Five (6) of the miRNAs are downregulated and the remaining 14 miRNAs are upregulated which is almost thrice the former. More so, the reports of upregulation of miRNAs (oncogenes) were triple times the report of downregulation (tumor suppressors).Table 3 also shows identified unvalidated 37 miRNA expressions in cervical cancer. These miRNAs are termed unvalidated because a single report of expression is not conclusive or affirmative of the diagnostic value of the miRNA. Ten (10) of the miRNAs are downregulated while 27 miRNAs are upregulated which is almost thrice the former. Table 4 shows that among the 66 miRNAs in cervical cancer identified in literature, 5 miRNAs grouped as Class I miRNAs (miR-21, miR-106a, miR-143, miR-145 and miR-203) had the highest number of reports in 6 to 5 research articles. Twelve miRNAs grouped as class II miRNAs were next to Class I in number of frequency (4 to 3 reports) while those miRNAs with less frequency (2 reports) were grouped as class III miRNAs. Class IV miRNAs are those ones with just a single report of identification in literature; there controversial status have not been determined yet.
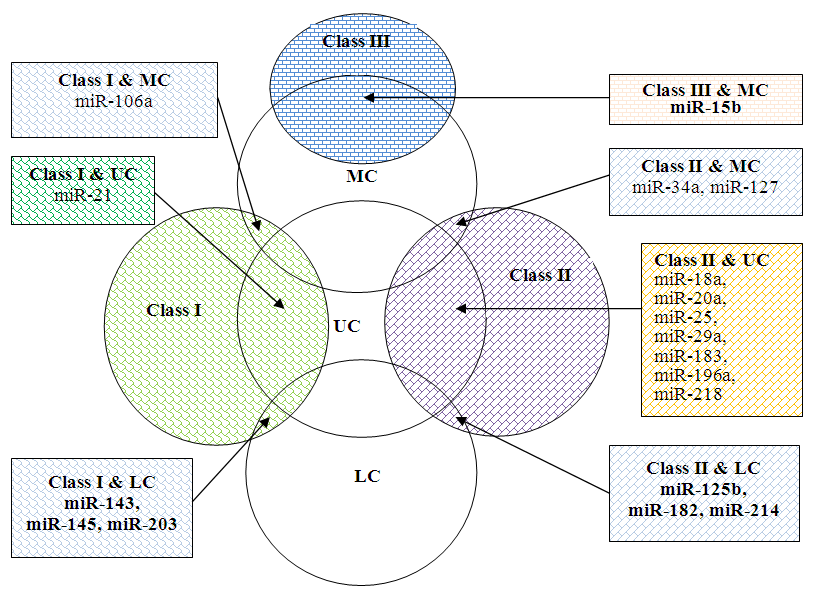 | Figure 1. Relationship between frequency of miRNA report and status of controversy |
|
|
|
|
4. Discussion
4.1. miRNA Expression in Human Papilloma Virus (HPV) and Herpes Simplex Virus (HSV) Infected Cervical Samples
- Almost all cervical cancers are caused by HPV infection; which is one of the major sexually transmitted infections [29-31]. HPV infects basal cells of the cervix through compromised architecture of the mucosa and skin due to sexual activity and is preserved as an episome. Once viral DNA is incorporated into host DNA, oncogenic transformation is induced [32]. An epidemiological study suggests that a biological interaction between HSV-2 and HPV-16 or HPV-18 occurs during the development of cervical carcinoma [33]. However, the interactions between HSV-2 and HPV-16 have not been fully elucidated. It is unclear whether such cervical carcinoma occurs due to direct virus-to-virus interaction, indirect immunologic response or oncogenic pathway. Several independent epidemiological studies have also linked HSV-2 infections to an increased incidence of cervical carcinoma [34]. A lot of reports have been made on how HPV through miRNA regulation trigger up cervical cancer development but little or no work has been carried out to evaluate HSV-miRNA regulatory effect in the emergence of cervical cancer. According to Liu et al. [35], three miRNAs (miR196a, 133a, and 133b) are significantly down-regulated in HPV16-positive normal cervix samples when compared with normal cervix samples. More so, Martinez et al. [11] reported that miR-182 and miR-183 are up-regulated while miR-1, miR-133b, miR-143, miR-145, miR-214, miR-368, miR-451, and miR-7029 are down-regulated in cell lines positive for HPV16 or HPV18 DNA. Yao et al. [36] revealed that HPV positive cells in uterine cervix harbor genes encoding miR-432, miR-1286, miR-641, miR-1290, miR-1287, and miR-95 which are hypermethylated while Wilting et al. [37] reported hypermethylation of miR-149, miR-203, and miR-375 in HPV-positive high-grade dysplasia and suggested that miR-203 and miR-375 may be markers for precancerous conditions in the uterine cervix. In a study conducted by Choi et al. [38], HSV induces Behçet’s Disease through upregulation of miR-21. Hence, the expression of miRNA in cervical cancer may not only be HPV dependent but could also be HSV-2 related. If that is the case, there is a possibility that the type of regulation (upregulation or downregulation) and degree of miRNA expression in a tissue infected with HPV alone may be different from that of HSV-2 and HPV co-infection. If this is true, it will go a long to explaining some of the controversy observed in table 1.
4.2. Differential Distribution of miRNAs on Solid Tumors
- According to the reports of Jian-Yi et al. [39], miRNA are differentially distributed on breast cancer tissues and the type of miRNA identified is dependent on the site (center to edge) of sample collection. They hypothesized that the differentially expressed miRNAs might be linked to the extent of hypoxia, or perhaps with angiogenesis. Their findings may provide some degree of explanation on the ambiguous miRNA regulation in cervical cancer as seen in some discordant reports (Table 1).
4.3. miRNA Expression in Cervical Cancers Stages
- The stages of cancer can be revealed by the level of miRNA expressions. Among other miRNAs, only miR-34 has shown the potential to classify cervical lesions. Here, the downregulation of miR-34a in CINI, CIN II, CIN III was observed to be +, ++ and +++, respectively when compared with normal tissue [15, 16]. The mechanism is linked to miR-34 downstream target of Bcl-2, Notch, and HMGA2 which are involved in cancer stem cell self-renewal and differentiation [40, 41]. This portrays miR-34 as a sensitive marker for cervical cancer classification. It was also reported that miR-218 in patients with CIN2 and CIN3 was lower than in patients with CINI [15]. According to Perieira et al. [19], the downregulation of miR-26a, miR-29a, miR-99a, miR-203, miR-513, miR-143 and miR-145 were all found to be equally downregulated in CIN, CINIII and Carcinoma while miR-10a, miR-196a and miR-132 were observed to be equally upregulated when compared with normal tissues, thus making them far from been used as specific quantitative differential indicators of cervical cancer stages. However, both the up and downregulated miRNAs can be assayed as complementary co-markers in qualitative CIN, CINIII and Carcinoma diagnosis. Some miRNAs can be used to differentiate certain cervical lesions from another as they show some degree of specificity and sensitivity both in the level and shift (direction) of expression. For example, miR-100 was less downregulated in CIN (+) when compare to Carcinoma (++) which is twice downregulated. More so, miR-148 was observed to be twice upregulated in CIN (++) when compared with Carcinoma (+++) which was trice upregulated [19, 14].
4.4. MiRNA Expression in Cervical Cancer Metastasis
- MiR-200a, miR-205 [42] and miR-127 [22] have been shown to regulate the metastatic potential of cancer cells to migrate to distant sites; hence, miR-200a could control cancer phenotype by regulating metastasis processes [26]. Furthermore, miR-20a upregulation level, as reported by Zhao et al. [43], correlates well with cervical cancer progression, particularly invasion and metastasis by targeting ATG7 and TIMP2. Other miRNAs associated with metastasis, progression, or survival of cervical cancer includes: miR-375 [44], miR-127 [22], miR-9 [26], miR-200a [26; 45], miR-93 [45], miR-497 [46], and miR-224 [47]. In the study carried out by Lee et al. [48], down-regulation of miR-494 is associated with poor patient survival, suggesting its possible role as a prognostic marker.
4.5. Chromosomal Instability
- Chromosomal instability, a characteristic of many solid tumors, is a state in which cells alter their chromosomal constituent at an increased rate [49, 50]. Its increase is directly proportional to increasing tumour grade and invasiveness [51-54]. A variety of mechanisms of chromosomal instability have been elucidated such as centrosome duplication [55-57], spindle assembly checkpoint defects [58, 59], telomere dysfunction [60], dysfunctional cell-cycle regulation [61-63], sister chromatid cohesion defects [64, 65], and control of adhesion of microtubules to chromosomes at the kinetochores [66-68]. According to How et al. [69], CIN70 (gene) score is significantly linked with chromosomal alterations, paraaortic distant relapse and patient overall survival in cervical cancer. It is logical to assume that there is a link between miRNA expression and Chromosomal instability in cervical cancer. It is also hypothetical that miRNA upregulation or downregulation in some tumor could be dependent on the particular mechanism initiating the chromosomal instability, rate of chromosomal alteration and target gene involved. Hence, the combined use of CIN70 signature and miRNA expression (especially miR-21) as co-markers in cervical cancer may offer better prognostic value in patient management.
4.6. MiRNA Controversy
- Considering the miRNA reports in table 1, more reports are in favour of downregulation of the identified 9 controversial miRNAs than upregulation, especially for miR-34a, miR-125b, miR-127, miR-143, miR-145, miR-203 and miR-214. The controversy with these miRNAs, especially miR-143, miR-145 and miR-203 may lie in the tendency of miRNA expression to shift to the left (that is, change course; downregulate from an upregulated expression). This scenario was observed with miR-522 which was twice upregulated in CIN (++) but singly (+) downregulated in Carcinoma (+) [14, 19]. The latter might serve as explanation for the controversy in table 1. Hence, the differences in miRNA expression among the reports of some researchers might be linked to the stage of cancer and perhaps the level of hypoxia or angiogenesis at the time of sample collection and analysis. Contrary to the reports of Wang et al. [13], Lui et al. [18], Martinez et al. [11] and Liu et al. [70] who observed that miR-143, miR-145 and miR-125b were highly upregulated in normal cervix, Lee et al. [22] and Dreher et al. [23] reported upregulation of the miRNAs but in cervical cancer. Thus, the controversy may lie in the nature (pathologic or non-pathologic state) of sample analyzed.High risk HPV exhibits their capabilities through three genes: E5, E6 and E7. MiR-196a is down-regulates in cervical cancer cell lines positive for HPV16 E5 but up-regulated in HPV16-negative cervical cancer cell lines. Furthermore, target gene HoxB8 is up-regulated in HPV16 cervical carcinoma cell lines but down-regulated in HPV18 cervical cell lines [35]. Contrary to the up-regulated miR-34a report of Martinez et al. [11] who worked on cervical cancer-derived Sitta and Caski cells, Chang et al. [71], He et al. [72] and Raver-Shapira et al. [73] reported down-regulation of miR-34a in cervical cancer positive for high-risk E6 oncoprotein. More so, among HPV16 and Epstein-Barr virus-derived cancers, miR-146a expression is apparently upregulated in cervical cancer [13], probably through NF-kappaB-dependent reaction [74-76]. However, miR-146a has been reported to be differentially downregulated in virus-independent cancers such as hormone refractory prostate cancer [77] and papillary thyroid carcinoma [78]. These events suggest that miRNA expression may be dependent on type of cell, viral agent and oncoprotein. Hence, the latter events may be one of the reasons for the observed controversies in table 1. Following the evaluation of miRNA expression in ovarian cancer, Resnick et al. [79] reported down-regulation of miR-155 in patient serum but Laios et al. [80] reported an up-regulation in primary ovarian tumours. Thus, some of the observed controversy in table 1 might be linked to the type of sample (serum vs tissue) used in assessing miRNA expression in cervical cancer. According to Lopez and Lopez [81], miR-7 is observed to be up-regulated in cancer when analyzed using microarray, cloning and sequencing technique but observed to be down-regulated when RT-PCR is used. In their report, miR-17-5p and miR-200c are down-regulated when investigated using RT-PCR but up-regulated when microarray technique is used while it is the reverse for miR-93 expression.Interestingly, there are more reports of upregulated miRNAs (especially the uncontroversial and unvalidated miRNAs) in literature than for down-regulated miRNAs (table 2 and table 3). This suggests that there are more oncogenic miRNA players in the initiation of cervical cancer than tumor suppressors. The most frequently reported miRNAs in cervical cancer includes: miR-21, miR-106a, miR-143, miR-145, miR-203 (table 4). Among these miRNAs, only miR-21 was found to be undoubtedly associated with cervical cancer. This could mean that miR-21 expression is unaffected by factors that bedevil other miRNAs. This finding suggests that miR-21 is a better prognostic marker in cervical cancer management. Be that as it may, in terms of frequency, there are other miRNAs that are moderately identified in cervical cancer which include: miR-18a, miR20a, miR-20b, miR-25, miR-34a, miR-125b, miR-127, miR-182, miR-183, miR-196a, miR-214, miR-218. However, only miR-18a, miR-20a, miR-25, miR-183, miR-196a, miR-218 were found to be uncontroversial (Figure 1). Hence, the latter miRNAs can specific be used as diagnostic markers of cervical cancer when miR-21 signature in tissues cannot be assessed.
5. Conclusions
- The controversy surrounding the expression of miRNAs in cervical cancer might be associated with factors such as stage of cancer, pathologic or non-pathologic state of the sample, technique employed, site of collection, type of sample (serum or tissue) and type of viral agent and oncoprotein present. Since miR-21 is more frequently reported without any discordance when compared with other miRNAs, it should be adopted as the best marker in monitoring and management of cervical cancer patients.
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML