-
Paper Information
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
Research In Cancer and Tumor
2013; 2(3): 49-56
doi:10.5923/j.rct.20130203.02
Significance of Telomerase Activity and Gene Expression in Colorectal Cancer
Ahu Izgi1, Armagan Gunal2, Ufuk Gunduz1
1Middle East Technical University, Department of Biological Sciences, Ankara, 06531, Turkey
2Gülhane Millitary Medical Academy, Department of Pathology, Ankara, 06010, Turkey
Correspondence to: Ahu Izgi, Middle East Technical University, Department of Biological Sciences, Ankara, 06531, Turkey.
| Email: |  |
Copyright © 2012 Scientific & Academic Publishing. All Rights Reserved.
Introduction: Telomerase is a ribonucleoprotein synthesizing telomeric DNA onto the chromosomes which has two functional subunits: hTERT (humantelomerasereversetranscriptase) and hTR (humantelomeraseRNA). Activation of telomerase appears to be associated with unlimited replicative potential. Telomerase activity and expression of telomerase components may have significant roles in diagnosis or/and prognosis of malignancies. PCR-based TRAP assay is very sensitive for the detection of enzymatic activity of telomerase even if a few number of cancerous cells are available. The diagnostic or prognostic value of telomerase activity and its expression in colorectal cancer are still unclear. MaterialandMethods:Protein extraction was performed from specimens of matched normal and colon cancer specimens. Protein concetrations were determined by Bradford assay. Optimized protein concentrations were used for TRAPAssay (Telomericrepeatamplificationprotocol) which is a PCR-based assay. TRAP products were seperated by vertical gel electrophoresis on 12.5% polyacrylamide gel and visualized by silver staining. Gene expression of hTERT was determined by qPCR analysis.Results:The results demonstrated that all tumor tissues were telomerase positive whereas all corresponding normal tissues were telomerase negative. Among clinicopathological findings, telomerase activity was found to be associated with stage, histology, localization, distant metastasis and lymph node metastasis of tumor. The expression of hTERT was observed both in tumor and normal tissues, however; tumor tissues expressed 4.33 fold more hTERT than normal tissues. Although all of the clinicopathological findings differed in the expression of hTERT compared to normal tissues, they did not differ from each other significantly.Conclusion:Telomerase activity could be a promising tumor marker for colorectal cancer and there is a close association between the enzymatic activity and the expression of its catalytic subunit.
Keywords: Telomerase, Colorectal Cancer, hTERT
Cite this paper: Ahu Izgi, Armagan Gunal, Ufuk Gunduz, Significance of Telomerase Activity and Gene Expression in Colorectal Cancer, Research In Cancer and Tumor, Vol. 2 No. 3, 2013, pp. 49-56. doi: 10.5923/j.rct.20130203.02.
Article Outline
1. Introduction
- Eukaryotic chromosomal ends are composed of tandemly repeated G-rich sequences which are also called as telomeres[1]. Telomeres protect the ends of chromosomes from degredation, end to end fusions and aberrant recombinations[2]. Normal somatic cells have limited proliferative capacity due to shortening of telomeres after each cell division[3]. Progressive shortening of chromosomal termini can result in loss of protective function of telomeres thereby loss of chromosomal stability and integrity[4]. Cells with critically shortened telomeres are recognized as a DNA damage response followed by growth arrest, therefore they go into replicative senescence[5]. In contrast to normal cells; stem cells, germline cells, hematopoietic cells, immortal cells and cancer cells are able to proliferate indefinetely thanks to the ability of stabilizing their telomeres by an enzyme called as telomerase preventing telomere loss[6,7]. Telomerase is a reverse transcriptase composed of two essential subunits: TERT (telomerase reverse transcriptase) and TR (telomerase RNA)[8]. Telomerase synthesizes hexameric telomeric repeats onto the chromosomes thereby compansating telomere loss in immortal cells such as tumor cells[9]. Telomerase utilizes TR subunit as a template since TR contains complemantary sequences to telomeric DNA sequences whereas TERT is used for nucleotide joining process as a catalytic subunit[10,11]. Activation of telomerase gene has been found in about %85 cancer types with the help of a sensitive PCR-based assay. Its activation leads to immortalization of cells through increasing proliferation capacity whereas most healthy normal tissues has shown little or no telomerase activity[12,13]. Its clinical importance varies among different types of cancer. Since there was no significant difference in telomerase activity between normal adjacent and tumor tissues, telomerase activity was found not to be a diagnostic or prognostic marker for some types of cancer whereas telomerase activity was found to be correlated with poor prognosis in certain types of cancer such as neuroblastoma, lung cancer, gastric cancer[14]. Previous studies demonstrated telomerase activity in colorectal cancer tissues and a few studies showed the expression level of hTERT in colorectal cancer together with telomerase activity[15-17]. However, the prognostic and/or diagnostic value of telomerase activity in colorectal cancer and its relation with clinicopathological features are still controversial. In our study, we analysed telomerase activity in both tumor and adjacent normal tissues by a quantitative telomeric repeat amplification protocol (TRAP)-silver staining assay. In addition, we also evaluated clinicopathological characteristics (such as age, gender, histological type, stage of disease, histological grade, tumor site, and size of tumors) regarding telomerase activity. Besides these criteria, depth of tumor, lymph node status and distant metastasis were also analysed. Moreover, the level of hTERT mRNA expression was analysed by quantitative- real time polymerase chain reaction (q-RT-PCR) both in normal tissue samples and tumor samples. We also evaluated clinicopathological findings with respect to the expression levels of hTERT.
2. Materials and Methods
2.1. Patients and Tissue Samples
- 20 colorectal cancer tissues and their adjacent normal tissues were obtained from patients at Gülhane Millitary Medical Academy (GATA) in Turkey. From each patient, 3 normal adjacent tissue sample and tumor tissue sample were obtained and studied. Ethical approval was obtained from the committee for the collection of tissue samples and further studies. Upon surgical removal, tumor and adjacent normal tissues were stored in RNAlater® solution immediately (Sigma-Aldrich, USA). After overnight incubation at 4°C, tissues were stored at -80°C. Additionally, main tissue specimens were fixed in 10% phosphate-buffered neutral formalin. Samples were embedded in parafin according to macroscopy guidelines. After 4µm sections were taken, slides stained with hematoxylin and eosin for histopathological analysis. Tumors were categorized in accordance with TNM system endorsed by American Joint Committee on Cancer (AJCC, Version7).
2.2. TRAP (Telomeric Repeat Amplification Protocol)-Silver Staining Assay
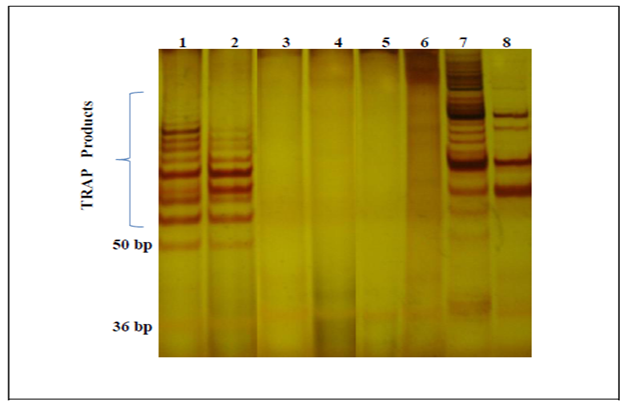
- Telomerase activity was determined in accordance with the telomeric repeat amplification protocol provided by the TRAPeze® telomerase detection kit (Millipore, Germany). Each sample was assayed in triplicate. Each experimental setup included a heat inactivated control, CHAPS lysis buffer only and a positive control. Fresh frozen tissue specimens were homogenized well with liquid nitrogen to a fine powder transferred with a sterile surgical blade to a RNAse free 1.5 mL microcentrifuge tube containing 40 unit/uL of RNAse inhibitor and 250 uL ice-cold CHAPS lysis buffer (10 mM Tris-HCl, pH 7.5, 1 mM MgCl2, 1 mM EGTA, 0.1 mM Benzamidine, 5 mM β-Mercaptoethanol, 0.5% CHAPS, 10% Glycerol). The homogenate was incubated on ice for 30 min and then spun in a benchtop microcentrifuge at 12.000 X g for 20 min at 4 °C. About 160 uL of supernatant was collected in a sterile RNAse free plastic tube, then stored at -80 °C for further use. Protein concentration of each sample was determined by Bradford assay. An aliquot of extract containing 0,6 ug of protein was used for each TRAP assay. PCR conditions performed were 30°C for 30 min for the elongation of TS ( telomere substrate) primer by telomerase and followed by 33 cycles of PCR amplification (94°C for 30 seconds, 59°C for 30 seconds and 72°C for 60 seconds). The PCR products were analyzed by vertical gel electrophoresis in a 12.5% polyacrylamide (PAGE) non-denaturating gel for 45 min at 300V. PAGE gel was stained with modified silver staining method. Specimens were considered telomerase positive showing six base pair incremental bands starting from 50 bp band according to manufacturers instructions. The gel images were analysed with Image J software. Relative telomerase activity (RTA) was calculated by the following formula;
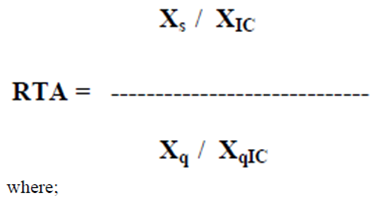

3. Reverse Transcription-quantitative Polymerase Chain Reaction (RT-qPCR)
3.1. RNA Isolation
- Total RNA was extracted from each tissue sample by using TRIzol Reagent according to manufacturers instructions (Sigma Aldrich, USA). The concentration and purity of isolated RNA samples were determined by measuring optical densities at 260 nm and 280 nm using NanoDrop 2000 C spectrophotometer (Thermo Fischer Scientific, USA). The intactness of isolated RNA and DNA contamination were checked by agarose gel electrophoresis.
3.2. cDNA Synthesis
- cDNA synthesis was performed by using QuantiTect Reverse Transcription Kit (Qiagen, Germany) according to manufacturers instructions. 1 ug total RNA and 25 pmol of gene specific primers, either of GAPDH, hTERT were used for each reaction. Briefly, genomic DNA elimination step was performed at 42°C for 2 min by using 2 ul genomic DNA wipeout buffer, 1 ug RNA to a final volume 11ul. Secondly, reverse transcription reaction was carried out at 42°C for 15 min by the addition of 4 ul RT buffer, 1 ul reverse primer or primer mix and 1 ul reverse transcriptase. Finally, the reverse transciption was terminated by incubation at 95°C for 3 min to inactivate reverse transcriptase. cDNA was stored at -20°C until further studies.
3.3. Real Time Quantitative Polymerase Chain Reaction
- hTERT mRNA levels were quantified by quantitative real time PCR (qRT-PCR) performed in Rotor-Gene 6000 (Corbett Research, Australia). Light-Cycler-FastStart SYBR Green I DNA master mix kit was used. Briefly, total volume of reaction mix was 20 ul containing 10 ul SYBR Green 2X master mix, 0.15 uM of reverse and forward primers, 2.8 uL of cDNA and appropriate amount of nuclease free water. All samples were run as triplicates in each run including a non-template control to check background signal. Relative quantification of qPCR products was perfomed by using 2-∆∆Ct method[18]. Delta delta Ct (2-DDCT) relative quantitation method was used for quantitation of qPCR products.
3.4. Statistical Analysis
- All data are representative of three independent experiments and expressed as mean _ standard error of the means (SEM). Relative telomerase activity (RTA) results of tumor and normal tissues were analyzed by two-tailed t tests to assess the association. The relationship between clinical variables and RTA values was analyzed by Mann-Whitney U test.The difference in the gene expression of hTERT between tumor and normal tissue samples was determined using t-test. One-way ANOVA test and post-hoc Tukey analyses were carried out to find groups whose mean differences were significant. The results were significant at the 0.05 level.
4. Results
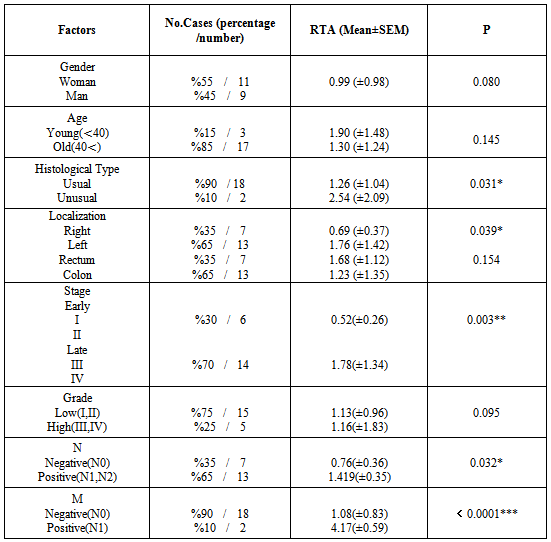
- 20 matched samples from patients with colorectal cancer were obtained and studied in this study. Tumors were localized mostly (13/20) in left colon and %65 of tumors in left colon were found in rectum and remaining were found in sigmoid colon. Tumors in right colon (7/20) localized in hepatic flexure, ascending colon, cecum. Tumor size ranged from 1.8 cm to 10 cm. According to histological analysis, 18 tumors out of 20 were usual adenocarcinomas whereas other two tumors were categorized as unusual adenocarcinoma which were both mucinous adenocarcinomas. According to TNM classification, 14 out of 20 patients were evaluated as late stage (stage III and IV) whereas 6 out of 20 patients were at early stage (stage I and stage II). One of the patient was diagnosed at early onset with Stage I which is a rare event in turkish population. About %65 of patients showed lymph node metastasis. The number of lymph node metastasis was variable, however; metastasis in 4 or more regional lymph node was common among patients evaluated. Distant metastasis (M) status was also evaluated and 2 out of 20 cases showed metastasis to liver and peritoneum. Patients with rectal tumors took chemoradiotheraphy as neoadjuvant therapy, however; other patients did not take any therapy before surgery.
4.1. TRAP-Silver Staining Assay
- In this study, all 20 of colorectal cancer cases showed telomerase activity at different levels, whereas none of the normal tissues were telomerase positive (Figure 1).
|
4.2. Expression Analysis of hTERT Gene in Colorectal Cancer Samples
- According to RT-qPCR results, all of the tumor and adjacent normal tissues were found to express hTERT, however; tumor tissues showed 4.33 fold more hTERT expression compared to their normal adjacent tissues (Figure 2A). Gene expression of hTERT was found to upregulate as cancer progressed, since tumors at late stages showed 3.13 fold more expression level of hTERT compared to normal tissue samples whereas tumors at early stage expressed high level of hTERT compared to normal tissues but statistically not significant (Figure 2B). The location (right and left colon), histology (usual and unusual types) and grade (II and III) of colorectal cancer tissues did not differ from each other significantly with respect to hTERT expression in the current study, but their expression was significantly higher than normal tissues (Figure 2C).
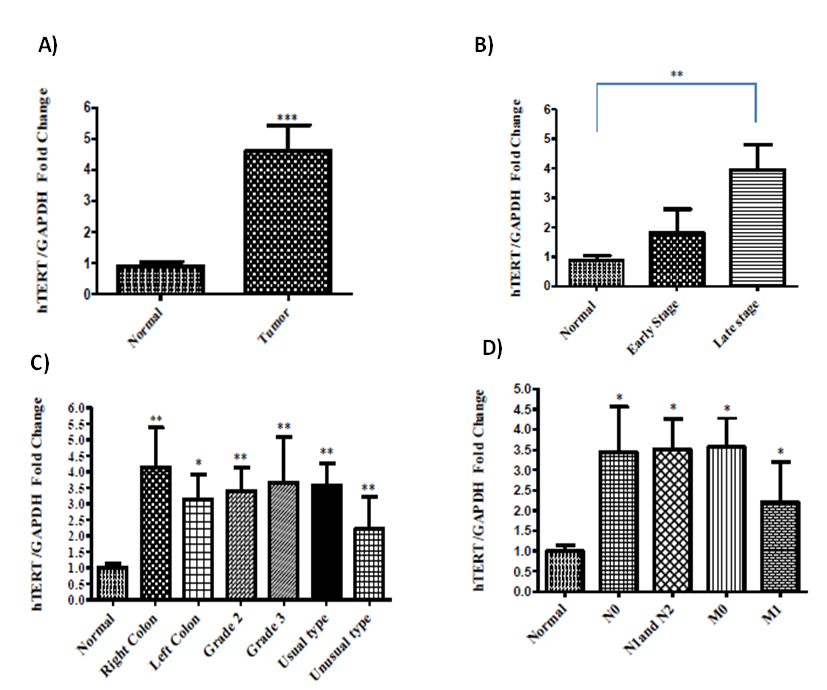 | Figure 2. hTERT expression levels in different clinicopathological parameters (* Results were signifcant with a p<0.05, ** results with a p < 0.01 and *** results with a p<0.0001) |
5. Discussion and Conclusions
- Maintenance of telomere length is prerequisite for cellular immortalization. Some cells including some type of tumors utilize different mechanisms other than telomerase activity for stabilizing and/or elongating of their telomeres, such as alternative lengthening of telomere (ALT) pathway, which is caused by homologous recombination or non homologous end-joining events between telomeric repeats[40]. Additionally, there are a number of telomere binding proteins which may have essential roles for maintanence of telomere length[41]. Although telomerase positive cells do not indicate necessarily malignancy, they are potentially immortal. On the other hand, telomerase activity was found to be strongly associated with the acquisition of malignancies and could be a potential biomarker for the detection of cancer[19]. Telomerase activity can be detected by four different methods which are TRAP assay, terminal rectriction fragment length analysis, Real-time PCR and immunohistochemistry. Each of these methods is used for detection of a different aspect of telomerase activity. Therefore, all four methods are not homogenous in research meaning[20]. TRAP assay is the best analysis method for enzymatic activity. RT-PCR has been used for the analysis of gene expression levels and it is a specific and sensitive method to analyse the expression level of hTERT which is the protein subunit of telomerase[21]. There are early studies that analysed telomerase enzyme activity in colorectal cancer; however, only a few studies conducted both in normal and cancerous tissues in colorectal cancer[22,23]. A variety of studies have investigated enzymatic activity of telomerase in colorectal cancer. However, whether the activity of telomerase in association with clinicopathological features of patients is still controversial[24-27]. Although some of the previous studies reported that normal colonic mucosa specimens showed telomerase activity, the current study demonstrated no telomerase activity in normal colonic mucosa specimens consistent with other studies which did not detect telomerase activity in normal colon tissues[28-30].Telomerase activity did not show any difference in age, gender in this study which is consistent with other studies; however, a northworty difference was observed between tumor site and telomerase activity. Tumors located at left colon had significantly higher telomerase activity compared to tumors at right colon. Previous study reported that proximal and distal colon carcinogenesis were found to have distinct biological characterictics which have been reported to be acquired during embryogenesis or postnatal development. Moreover, it was stated that different procarcinogenic pathways are responsible for the development of carcinogenesis in right and left colon which may be related with the difference in the relative telomerase activity between right and left colon[31]. In the current study, telomerase activity was found to increase with the stage of the tumor which is in consistence with previous study[32]. Telomerase activity could be an indicator of agressiveness of malignancies[33]. Among histopathological findings, the unusual histological type, mucinous adenocarcinomas, showed significantly higher telomerase activity compared to usual colorectal adenocarcinomas in this study. Moreover, of the TNM classification parameters, metastasis (M) and lymph node metastasis (N) positive tumors demonstrated higher telomerase activity in the current study. High telomerase activity may accelerate abnormal cell proliferation and help tumor cells escape from apoptosis due to shortened telomeres[34]. In this study, there is only one tumor sample was determined with pT1; therefore, statistical analysis between RTA and depth of tumor could not be perfomed. Similarly, Shoji et. al, found a relationship between telomerase activity and depth of invasion of tumors (pT)[35]. In two cases out of twenty of the current study, liver metastasis was observed with high telomerase activity. A variety of studies conducted so far revealed that hTERT could serve as a potential and useful tumor marker thanks to its expression in a number of different cancer types. Therefore, hTERT could be used as a screening tool for diagnostic or prognostic purposes by real-time PCR[36,42]. In the current study, although all tumor and normal tissue specimens were found to express hTERT, tumor specimens expressed more hTERT compared to normal tissues which is in accordance with previous study conducted in colorectal cancer[36]. Simirlary, in this study telomerase activity was found to have a tendency to be higher in tumor tissues compared to corresponding normal tissues. As cancer progressed, hTERT expression was found to be increased. Late stage tumors expressed more hTERT than early stage tumors and normal tissues. Likewise, late stage tumors showed higher telomerase activity compared to early stage tumors. These results showed there could be a close correlation between telomerase activity and hTERT expression. However, other clinicopathological findings did not differ from each other in the expression level of hTERT in this study whereas some of previous studies reported a correlation of telomerase catalytic subunit expression with depth of tumor and tumor grade[36,37]. Some of earlier results were consistent with the results of current study which were unable to demonstrate a correlation among the expression of telomerase catalytic subunit and clinicopathological characteristics of tumors such as tumor site, lymph node involvement and distant metastasis[36]. In contrast, a study revealed a significant difference between expression of hTERT and tumor site. Moreover, another study demonstrated that hTERT expression correlated with tumor grade[37]. In the current study, normal tissues were telomerase negative; however, they were shown to express hTERT gene. This may be due to posttranscriptional splicing of hTERT or posttranslational regulation of hTERT gene. Kilian et.al 1997, reported that posttranscriptional splicing of hTERT products may lead to production of different proteins with different functions[38]. Alternatively, alternative splicing of hTERT variants may cause the deletion in the protein structure[39].The absence of telomerase activity in normal adjacent tissues may indicate that the enzymatic activity of telomerase and hTERT expression may be clinically useful as a diagnostic or prognostic tool. Moreover, the activity of telomerase and its catalytic subunit expression was found to correlate significanlty in colorectal cancer in the current study. These results may indicate that telomerase could be a promosing marker for colorectal cancer in the future. In this study, some relations or and associations found was statistically non-significant. This could be related with a limited number of patients categorized in different clinicopathological parameters. Thus, it may be useful to demonstrate these results in a larger number of patients’ pool.
References
| [1] | van Steensel B, Smogorzewska A. & de Lange, T. TRF2 protects human telomeres from end‑to‑end fusions. Cell 1998; 92, 401–413. |
| [2] | Hastie ND, Dempster M, Dunlop MG, Thompson AM, Green DK, Allshire RC. Telomere reduction in human colorectal carcinoma and with ageing. Nature 1990 Aug 30;346 (6287):866-8. |
| [3] | Watson J. D."Origin of concatemeric T7 DNA." Nat New Biol.1992; 239(94):197-201. |
| [4] | Counter CM, Avilion AA, LeFeuvre CE, Stewart NG, Greider CE, Harley CB and Bacchetti S. Telomere shortening associated with chromosome instability is arrested in immortal cells which express telomerase activity. EMBO J. 1992 May; 11(5): 1921–1929 |
| [5] | Chen JH, Hales CN, Ozanne SE. DNA damage, cellular senescence and organismal ageing: casual or correlative. Nucleic Acids Res. 2007; 35(22): 7417–7428. |
| [6] | Wright WE and Shay JW. The two-stage mechanism controlling cellular senescence and immortalization. Exp. Gerontol. 1992; 27,383-389 |
| [7] | Brümmendorf TH. Telomerase Activity- a Prognostic Factor in Colorectal Cancer. Onkologie 2005; 28: 550-551 |
| [8] | Theimer C& Feigon J. Structure and function of telomerase RNA. Current opinion in structural biology 2006; 16(3), 307-18. |
| [9] | Blackburn EH. Telomere structure and synthesis. J Biol Chem.1990; 265, 5919-5921. |
| [10] | Nakamura TM, Morin GB, Chapman KB, et al. Telomerase catalytic subunit homologs from fission yeast and human. Science 1997;277:955–9. |
| [11] | Autexier C, & Lue NF. The Structure and Function of Telomerase Reverse Transcriptase. Annu Rev Biochem. 2006;(75), 493-517. |
| [12] | Meeker AK, Coffey DS. Telomerase: a promising marker of biological immortality of germ, stem and cancer cells. A review, Biochemistry 1997; 62, 1323-31 |
| [13] | Kim NW, Piatyszek M, Prowse KR, Harley CB, West MD. Ho PL, Coviello GM., et al. Specific association of human telomerase activity with immortal cells and cancer. Science 1994; 266 (5193), 2011-2015 |
| [14] | Hiyama E, Hiyama K, Yokoyama T, Matsuura Y, Piatyszek MA, Shay JW. Correlating telomerase activity levels with human neuroblastoma outcomes. Nat Med. 1995; 1(3):249-55. |
| [15] | Ghori A, Usselmann B, Ferryman S, Morris A, Fraser I. Telomerase expression of malignant epithelial cells correlates with Dukes' stage in colorectal cancer. Colorectal Disease 2000; 4(6), 441–446. |
| [16] | Kawanishi-Tabata, R Lopez F, Fratantonio S, Kim N, Goldblum J, Tubbs R, Elson P, et al. Telomerase activity in stage II colorectal carcinoma. Cancer 2002; 95(9), 1834-9. |
| [17] | Roig AL, Wright WE, & Shay JW. Is Telomerase a Novel Target for Metastatic Colon Cancer, Current Colorectal Cancer Reports 2009; 5: 203–208 |
| [18] | Livak KJ, Schmitthen TD. Analysis of relative gene expression data using realtime quantitative PCR and the 2(Delta Delta C(T)) Methods 2001;25(4), 402-408. |
| [19] | Chadeneau C, Hay K , Hirte H, Gallinger S, Bacchetti S. (1995) Advances in Brief Telomerase Activity Associated with Acquisition of Malignancy in Human Colorectal Cancer. Medicine, 2533-2536. |
| [20] | Smith R, Lam A, K-Y Telomerase Activity: Detection and Clinical Implications in human cancers. Clinical oncology and search 2009; 2009-24614. |
| [21] | Bustin SA. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J Mol Endocrinol. 2000; 25: 169-193. |
| [22] | Fang DC, Joanne Y, Luo Y, et al. Detection of telomerase activity in biopsy samples of colorectal cancer. J Gastroenterol Hepatol. 1999; 14, 328-332. |
| [23] | Yan P, Saraga EP, Bouzourene H, et al. Telomerase activation in colorectal cacinogenesis. J Pathol.1999; 189: 207-12. |
| [24] | Hiyama E, Hiyama K, Yokoyama T, Matsuura Y, Piatyszek M.A and ShaJ W. Correlating telomerase activity levels with human neuroblastoma outcomes. Nat. Med.1995; (1), 249-255 |
| [25] | Hiyama E,Yokoyama T,Tatsumoto N, Hiyama K, Imamura Y,Murakami Y, Kodama T, Piatyszek MA, Shay JW, Matsuura Y.Telomerase activity in gastric cancer. Cancer Res.1995; (55), 3258-3262. |
| [26] | Kyo S, Kanaya T, Ishikawa H, Ueno H, Inoue M. Telomerase activity in gynecological tumors. Clin. Cancer Res. 1996; (2); 2023-2028. |
| [27] | Roig AI, Wright, W. E, & Shay J W. Is Telomerase a Novel Target for Metastatic Colon Cancer, Current Colorectal Cancer Reports 2009; 5, 203–208. |
| [28] | Ghori A, Usselmann B, Ferryman S, Morris A, Fraser I.(2000). Telomerase expression of malignant epithelial cells correlates with Dukes' stage in colorectal cancer. Colorectal Disease 4(6), 441–446. |
| [29] | Yan, P, & Bosman, F. T. (1998). Diagnostic Techniques Tissue Quality is an Important Determinant of Telomerase Activity as Measured by TRAP Assay. Biotechniques, 25(4), 660-662. |
| [30] | Tang R, Cheng, a J, Wang, J. Y& Wang T C. (1998). Close correlation between telomerase expression and adenomatous polyp progression in multistep colorectal carcinogenesis. Cancer research, 58(18), 4052-4. |
| [31] | Glebov OK, Rodriguez LM, Nakahara K, et al. Distinguishing right from left colon by the pattern of gene expression. Cancer Epidemiol Biomarkers Prev 2003; 12:755–62. |
| [32] | Engelhardt M, Drullinsky P, Guillem J, & Moore Ma. (1997). Telomerase and telomere length in the development and progression of premalignant lesions to colorectal cancer. Clinical cancer research: an official journal of the American Association for Cancer Research, 3(11), 1931-41. |
| [33] | Ghori A, Usselmann B, Ferryman S, Morris A, Fraser I.(2000). Telomerase expression of malignant epithelial cells correlates with Dukes' stage in colorectal cancer. Colorectal Disease 4(6), 441–446. |
| [34] | Saito Y, Kosugi S, Wakabayashi Y, Mishima Y, Hatakeyama K, Kominami R (1997)Telomerase activity and metastasis: expansion of cells having higher telomerase activity within culture lines and tumor tissues, Jpn J Cancer Res, 88(8)-732-7. |
| [35] | Shoji Y, Yoshinaga K, Inoue A, Iwasaki A, Sugihara K 2000. Quantification of telomerase activity in sporadic colorectal carcinoma. Cancer 88(6), 1304–1309. |
| [36] | Gertler R, Rosenberg R, Stricker D, Werner M, Lassmann S, Ulm K, Nekarda H, et al. (2002). Prognostic potential of the telomerase subunit human telomerase reverse transcriptase in tumor tissue and nontumorous mucosa from patients with colorectal carcinoma. Cancer 95(10), 2103-11. |
| [37] | Terrin L, Rampazzo E, Pucciarelli S, Agostini M, Bertorelle R, Esposito G, DelBianco P, et al. (2008). Relationship between tumor and plasma levels of hTERT mRNA in patients with colorectal cancer: implications for monitoring of neoplastic disease. Clinical cancer research: an official journal of the American Association for Cancer Research, 14(22), 7444-51. |
| [38] | Kilian A, Bowtell D.D.L. and Abud H.E. (1997) Isolation of a candidate human telomerase catalytic subunit gene, which reveals complex slicing patterns in different cell types. Hum. Mol. Genet., 6, 2011–2019. |
| [39] | Yi X, White D.M, Aisner D.L, Baur J.A, Wright W.E, Shay J.W (2000) An Alternate Splicing Variant of the Human Telomerase Catalytic Subunit Inhibits Telomerase Activity. Neoplasia 2(5): 433–440. |
| [40] | Reddel RR, Bryan TM, Colgin LM, Perrem KT and Yeager TR. Alternative lengthening of telomeres in human cells. Radiat Res. 2001; 155: 194-200. |
| [41] | Zhong Z, Shiue L, Kaplan S & de Lange, T. (1992). A mammalian factor that binds telomeric TTAGGG repeats in vitro. Mol. Cell. Biol. 13, 4834-4844. |
| [42] | Kirkpatrick KL, Mokbel K.(2001). The significance of human telomerase reverse transcriptase (hTERT).EJSO. 27: 754-760. |
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML