-
Paper Information
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
International Journal of Plant Research
p-ISSN: 2163-2596 e-ISSN: 2163-260X
2017; 7(4): 90-93
doi:10.5923/j.plant.20170704.02

Genetic Analysis of F2 Population of Tomato for Studying Quantitative Traits in the cross between Coldera x KHT5
Bilal Ahmed Khan1, Syeda Faiza Mehboob2, Mehboob Ahmad1, Mazhar Iqbal1, Ihsan Ullah1, Maria Saleem1, Adil Rehman3, Muhammad Shaid3
1Hazara Agricultural Research Station, Abbottabad, Pakistan
2Hazara University, Mansehra, Pakistan
3Agricultural Research Station, Mansehra, Pakistan
Correspondence to: Bilal Ahmed Khan, Hazara Agricultural Research Station, Abbottabad, Pakistan.
| Email: |  |
Copyright © 2017 Scientific & Academic Publishing. All Rights Reserved.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/

F2 population of cross Wild tomato (Solanum pimplifollium) variety Cauoldera (Wild) and cultivated tomato (Solanum lycopercicum) KHT5 was evaluated to find genetic components i.e., genotypic variance (Vg), phenotypic variance (Vp), genotypic coefficient of variance (GCV), phenotypic coefficient of variance (PCV), heritability (Hb) and genetic advance (GA) for quantitative characteristics of tomato. Data were recorded for P1, P2, F1 and F2 population for flower cluster-1, fruit cluster-1, fruit weight, fruit diameter, plant height, fruit plant-1 and yield plant-1, respectively. All traits revealed higher phenotypic and lower genotypic variances. Relatively, higher differences were recorded between phenotypic and genotypic coefficient of variance for all characteristics except fruit diameter. Maximum (181.66 & 104.15) and minimum (2.85 & 15.65) genotypic and phenotypic coefficient of variance was recorded for traits i.e., yield plant-1, plant height, flower cluster-1 and fruit diameter, respectively. Majority of the traits i.e., plant height, fruit diameter, yield plant-1 and fruit cluster-1 revealed highest values of heritability and genetic advance in means. Highest and lowest (99.83% & 24.56%) values of broad sense heritability with maximum and minimum (181.84% & 38.88%) values of genetic advance was recorded for traits i.e., plant height, fruit weight and flower cluster-1, respectively.
Keywords: Tomato, F2, Genetic analysis, Heritability, Genetic advance
Cite this paper: Bilal Ahmed Khan, Syeda Faiza Mehboob, Mehboob Ahmad, Mazhar Iqbal, Ihsan Ullah, Maria Saleem, Adil Rehman, Muhammad Shaid, Genetic Analysis of F2 Population of Tomato for Studying Quantitative Traits in the cross between Coldera x KHT5, International Journal of Plant Research, Vol. 7 No. 4, 2017, pp. 90-93. doi: 10.5923/j.plant.20170704.02.
Article Outline
1. Introduction
- Tomato (Solanum lycopersicum) is an herbaceous, usually sprawling plant in the order solanales and nightshade family, Solanaceae. It is an enrich source of vitamin C, lycopene and acts as powerful antioxidant and prevents cancer. It is the second most cultivated specie after potato due to its high nutritional values, cooking demand and wide range of value added products i.e., ketchups, sauces, pastes, soups, juices and purees. Intense need is being felt to increase the production of this crop due to increasing world population and consequently increasing consumption (Ahmad et al., 2015).Tomato is a unique vegetable crop, which is highly amenable to genetic improvement owing to its high degree of homogeneity and ease of controlled pollination (Pradeepkumar et al., 2001). The development of an effective plant breeding programme is depending upon the assessment of polygenic variation, selection of elite genotypes, choice of parents and breeding procedures. Crop improvement depends upon the magnitude of genetic variability and the extent to which desirable characters are heritable. Genetic variability for yield and yield components is essential in the base population for successful crop improvement (CR. W. Allard, 1960). Assessment of genetic variation and degree of transmission of desirable characters is helpful for planning a sound breeding programme (Shashikanth et al., 2010).The genetic variance of any quantitative trait is composed of additive variance (heritable) and non-additive variance and includes dominance and epistasis (non-allelic interaction). Therefore, it becomes necessary to partition the observed phenotypic variability into its heritable and non-heritable components with suitable parameters such as phenotypic and genotypic coefficient of variation, heritability and genetic advance (Mohammad et al., 2012). Plant breeders are continuously endeavouring to improve the genetic potential of yield and quality traits of tomato crop so as to meet the demands of an ever-increasing population of the world. Heritability determines how much of the phenotypic variability has a genetic origin and how much due to influence of environment, and therefore helps us select on genetic basis (Falconer 1981). Genetic advance is another parameter on which effectiveness of selection depends (Johnson et al. 1955). For the selection to be effective and for making improvement in the crop, moderate or high heritability should be accompanied by sufficient amount of genetic advance (Eid 2009). The approaches to make significant improvement in tomato production require information regarding nature and magnitude of genetic variation in quantitative traits and their interrelationships in the available germplasm, which are important pre-requisites for a systematic breeding program (Firas et al., 2012). Researchers have emphasized on estimation of genetic components such as coefficient of variation, heritability and expected genetic advance in the prediction of response quantitative traits to selection (Mohamed et al., 2012; Dar and Sharma, 2011; Saeed et al., 2007; Mohanty et al., 2003; Mohanty et al., 2002). The current research has been carried out to find the variance components like genotypic variance (Vg), phenotypic variance (Vp), genotypic co-efficient of variance (GCV), phenotypic coefficient of variance (PCV), heritability and genetic advance for yield and yield components. The study will be helpful in making desirable selection in F2 generation for the desired parameters in future breeding program.
2. Method and Materials
2.1. Field Data
- The current study was carried out at Hazara Agricultural Research Station, Abbottabad during the sowing seasons 2014 to 2016. Two varieties of diverse characteristics i.e KHT-5 (Solanum Lycopercicum with oval shaped fruit) and Coldera (Solanum pimpillifoilum) were crossed by determining KHT-5 as line and Coldera as tester. Seed was collected from the fruits obtained by the crossing. The collected seed was sown as F1during the months of April and transplanted in June 2015 and F2 seed was collected for F2 generation after proper compilation and analysing of F1 recorded data. During January 2016, F2 population was sown in nursery and then transplanted into the field in the month of March, by keeping 50 cm plant to plant and 100 cm row to row distance. Data were collected on all the plants in F2 population at appropriate time, using protocol described for each trait i.e., number of flowers per cluster, Number of fruits per cluster, Fruit weight, Fruit diameter, Plant height, Number of fruits per plant and Yield per plant.
2.2. Statistical Analysis
- Phenotypic (PCV) and genotypic (GCV) coefficients of variation were calculated according to the following formula proposed by Singh and Chaudhary 1985.
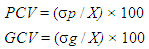 Where,σ p, σ g, and X are the phenotypic, genotypic standard deviation and grand mean of the traits, respectively.Vg (Variance of genotype), Vp (variance of phenotype) and broad sense heritability (Hb) were calculated on MS-excel by using the following formula suggested by Globerson et al. 1987.
Where,σ p, σ g, and X are the phenotypic, genotypic standard deviation and grand mean of the traits, respectively.Vg (Variance of genotype), Vp (variance of phenotype) and broad sense heritability (Hb) were calculated on MS-excel by using the following formula suggested by Globerson et al. 1987.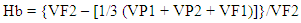 Where,
Where,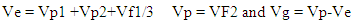 So, Hb = Vg/VpExpected genetic advance (GA) and percentage of GA were calculated according to following formula proposed by Shukla et al. 2006.Expected genetic Advance (GA)
So, Hb = Vg/VpExpected genetic advance (GA) and percentage of GA were calculated according to following formula proposed by Shukla et al. 2006.Expected genetic Advance (GA) 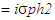 Percentage of Genetic Advance (GA %)
Percentage of Genetic Advance (GA %) 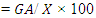 Where, I: standardized selection differential, a constant (2.06), σp: phenotypic standard deviation.
Where, I: standardized selection differential, a constant (2.06), σp: phenotypic standard deviation.3. Results
- The results for Phenotypic variance (Vp), Genotypic variance (Vg), Genotypic Co-efficient of variance (GCV), Phenotypic Co-efficient of Variance (PCV), Heritability% (Hb) and Genetic Advance % (GA%) in F2 population of cross combination Coldera × KHT5 are shown in Table-1. Analyzed data revealed that genotypic variance was found lower than phenotypic variance for all the traits under observation. Further, higher difference was observed between genotypic and phenotypic variances for the traits i.e., Fruit weight, fruits plant-1 and yield plant-1 while lower difference was found for Flowers cluster-1, fruit cluster-1, fruit diameter and plant height (Table 1).Relatively higher difference was noted between genotypic and phenotypic co-efficient of variances for the traits i.e., Flowers cluster-1, fruits cluster-1, fruit weight, plant height, fruit plant-1 and yield plant-1 while fruit diameter showed lower differences for GCV and PCV. Highest value of PCV (104.15%) and GCV (181.66%) were observed in the traits i.e., plant height and yield plant-1, respectively which indicates that these traits has highest diversity means, so, there is wide range of selection for the breeder. Lowest values of PCV (15.65%) and GCV (2.85%) were recorded for fruit diameter and flower cluster-1, respectively means variation in the trait is lowest than in any other trait so range for selection is narrow (Table 1).Maximum (99.83%) broad sense heritability % was observed in plant height followed by fruit diameter and yield plant-1 having broad sense heritability values of 99.70 % and 92.68%, respectively which reveals that strong additive gene action with fewer chances of environmental effects. Minimum (24.56%) broad sense heritability was recorded for the fruits weight while moderate (49.41%) broad sense heritability was shown by flower cluster-1 (Table 1).
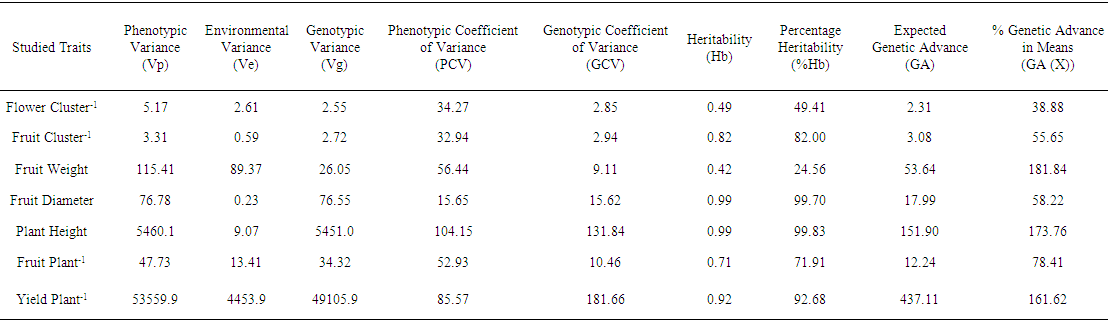 | Table 1 |
4. Discussion
- Data regarding the difference between variance of genotypes (Vg) and variance of Phenotypes (Vp) reveal that our findings matched to the study of Mohamed et al. 2012 and Gosh et al. 2010 for the traits i.e., fruit weight, fruit per plant and fruit yield per plant while others traits like flower per cluster, fruit per cluster, fruit diameter and plant height showed negligible differences between Vg and Vp indicating that they were less influenced to environmental factors for their phenotypic expression that might be due to strong genetic diversity existence between traits.High PCV reported for the traits: fruit per cluster, fruit weight and fruit per plant are similar with the results of Mohamed et al. 2012, Taina et al. 2015 and Kumar et al. 2001. Kaushik et al. 2011 also found small difference between GCV and PCV for fruit diameter. Sivaprasad et al. 2009 and Ahmad et al. 2015 reported high value of GCV and PCV for yield per plant which is in accordance with our findings. High GCV was observed for plant height similar with the findings of Saleem et al. 2013, Shanker et al. 2013 and Tiana et al. 2015. High PCV and GCV indicated high scope for selection and improvement in future.Lower difference between GCV and PCV for fruit diameter and plant height reports that strong genotypic interaction has found with fewer environmental effects while higher difference between GCV and PCV for other traits revealing presence of dominance and non-additive gene actions by showing maximum influence of environmental effect.Our result regarding number of fruits per cluster, fruit diameter, plant height and yield per plant tallies with the findings of Shashikanth et al. 2010, Shanker et al. 2013, Ready et al. 2013, Muhammad et al. 2012 and Gosh et al. 2010, respectively as they also found high value of heritability and GA% for the respective traits. It has indicated the potentiality of this cross combination to release variability and selection for these characters would give good response and the plant material for other characters can be improved through hybridization and selective breeding.Gosh et al. 2015 found relatively lower value of GA% (38.88%) coupled with low value of heritability % for flower per cluster which is similar to our findings for the respective trait. High genetic advance with moderate and low heritability by the trait i.e., fruit per plant and fruit weight were found which are in accordance to the findings of Gosh et al 2012, Ready et al. 2012, Shashikanth et al. 2010, Tianna et al. 2015 and Kumar et al. 2001, respectively. High heritability with high genetic advance for the traits are deemed to be under control of additive genes, whereas with high heritability and low genetic advance are under the control of non-additive (dominant and/or epistatic) genes which limits the scope of improvement through selection (Akbar et al. 2003).
5. Conclusions
- Analyzed data showed that variance of genotype was lower than the variance of phenotype in the F2 populations. Higher differences was observed for traits i.e., Flowers per cluster, fruit weight, fruit per plant and yield per plant while other traits revealed lower differences. Relatively higher differences were recorded between phenotypic and genotypic co-efficient of variances for the traits i.e., Flowers per cluster, fruits per cluster, fruit weight, plant height, fruit per plant and plant yield. The heritability ranged was 24.56% to 99.83%, as, majority of the traits showed highest values of heritability. The genetic advance ranged was 2.31 to 437.11. Highest and lowest genetic advance was found in yield per plant and flower per cluster, respectively. It indicates that we have wide range for selection in F2 population and these traits can further be exploited by direct selection for improving yield in tomato.
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML