-
Paper Information
- Next Paper
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
International Journal of Plant Research
p-ISSN: 2163-2596 e-ISSN: 2163-260X
2015; 5(1): 7-12
doi:10.5923/j.plant.20150501.02
Population Genetic Study of Eggplants (Solanum) Species in Nigeria, Tropical West Africa, Using Molecular Markers
C. U. Aguoru 1, L. O. Omoigui 2, J. O. Olasan 1
1Department of Biological Sciences, University of Agriculture, Makurdi, Nigeria
2Department of Plant breeding and Seed Science, University of Agriculture, Makurdi, Nigeria
Correspondence to: C. U. Aguoru , Department of Biological Sciences, University of Agriculture, Makurdi, Nigeria.
| Email: |  |
Copyright © 2015 Scientific & Academic Publishing. All Rights Reserved.
Population genetics of eggplants (Solanum species) was studied using RAPD (Random Amplified Polymorphic DNA) markers in six states (populations) within Nigeria, Tropical West Africa. The aim was to estimate the actual amount of polymorphism in each population and the overall population combined together. Binary matrices were manually computed from amplified DNA and resolved DNA bands. Statistical operation was performed using POPGEN32 software (1.31 versions) to generate population genetic parameters and dendrogram. Results showed that the mean effective number of alleles for Nasarawa State, FCT, Niger State, and Plateau State are 1.542, 1.137, 1.361 and 1.425 respectively. Kogi and Benue States had a close value of 1.2912 and 1.255 respectively. The number of polymorphic loci among the eggplants ranged from 5 in FCT to 19 in Nasarawa State. Nasarawa State had the highest percentage polymorphism of 63.3% followed by Plateau State (53.3%) and Niger State (43.3%). Kogi and Benue States recorded the same polymorphism of 40% while FCT was the lowest (16.67%). However, the multiple population Genic Variation Statistics (GVS) showed a total number of 25 polymorphic loci among the eggplants with a combined polymorphism of 83.33%. Benue and Niger States had the lowest genetic distance of 1.56 which was very far from FCT that recorded the highest distance of 7.016. Plateau and Nasarawa States had a genetic distance of 5.45 and 5.91 respectively. Consequently, Nasarawa state as a region of eggplant diversity could be explored by plant breeders and taxonomists while the rare genotypes of FCT, Benue state and Kogi states require urgent conservation measures as well as intensive cultivation of varied species.
Keywords: Eggplant, Genetic diversity, RAPD, Genetic distance, Conservation
Cite this paper: C. U. Aguoru , L. O. Omoigui , J. O. Olasan , Population Genetic Study of Eggplants (Solanum) Species in Nigeria, Tropical West Africa, Using Molecular Markers, International Journal of Plant Research, Vol. 5 No. 1, 2015, pp. 7-12. doi: 10.5923/j.plant.20150501.02.
1. Introduction
- Eggplants (Solanum species) belong to the family Solanaceae and consist of about 1500 morphologically diverse species (Sharmin et al., 2011; Sifau et al., 2014). Basically, among the commonly reported species included: S.melongena, S.macrocarpon, S.aethiopicum, S.incanum, S.scabrum, S.dasyphyllum, S.erianthum (Agnieszka et al. 2007; Osei et al., 2010; Knapp et al., 2013; Mariola et al., 2014; Sifau et al., 2014). Eggplant, a vegetable and fruit crop is intensively cultivated in many communities in Nigeria and many parts of the world where it is known for its nutritive, medicinal and other ethnobotanical uses (Ripperger and Himmelreich, 1994; Bello et al., 2005; Schippers, 2000; Shalom et al., 2011; Ali et al., 2013; FAOSTAT, 2014). International Plant Genetic Research Institute (IPGRI) (2001) emphasized on the need to detect polymorphisms among cultivars and lines for a successful breeding programme. This pool of genetic variation within an inter-mating population is the basis for selection as well as for plant improvement. Thus, conservation of this plant genetic diversity is essential for present and future human well-being ((IPGRI, 2001; Asthana and Asthana, 2012). During recent years there has been increasing awareness of the importance of adopting a holistic view of biodiversity including agricultural biodiversity and conservation for sustainable utilization and development. These principles have been enshrined in the convention on biological diversity and the global plan of action of the Food and Agricultural Organization of the United Nations (FAO, 2000). The emphasis is now to understand the distribution and extent of genetic diversity available to humans in plant species so that the genetic diversity can be safely conserved and efficiently used. Domestication, mutation, natural intercrossing, genetic drift, gene flow, selected breeding and hybridization have brought extensive genetic diversity in crops grown across different populations all over the world (Taylor et al., 2007). It is known that members of populations can interbreed and exchange their genes (gene flow).Therefore, gene pool system, the total variety of alleles and genes in a sexually reproducing population, is dynamic. A population whose gene pool shows constant change from generation to generation is actually undergoing evolutionary change leading to continuous process of speciation. The number of organisms in a population carrying a particular allele determines the allele frequency of that population and this frequency is useful in calculating the genotype frequency through a mathematical relationship developed by G.H. Hardy and W. Weinberg in 1908 called Hardy-Weinberg relationship (Taylor et al. 2007).Much of the large amount of diversity of a species may be found within individual populations or partition among a number of different populations (Taylor et al., 2007). A better understanding of genetic diversity and its distribution is essential for its conservation and use. It will help us in determining what to conserve as well as where to conserve and will improve our understanding of the taxonomy and origin and evolution of plant species of interest (IPGRI, 2001). Over the years across different parts of the world, studies on eggplant had concentrated on the taxonomic characterization of the crop using morphological, cytosytematic, phytochemical and molecular characterization (Khorsheduzzaman et al. 2008; Osei et al., 2010; Shalom et al., 2011; Hurtado et al., 2012; Fabio and Sabatino, 2013; Mariola et al., 2014). However, information on the population genic variations of the crop is limited. This study therefore emanated from the need to study the genetic diversity of eggplants across populations in Nigeria for the purpose of sustainability and breeding programmes. The aim was to estimate the actual amount of polymorphisms in six states of Nigeria using RAPD markers as a molecular approach.
2. Materials and Methods
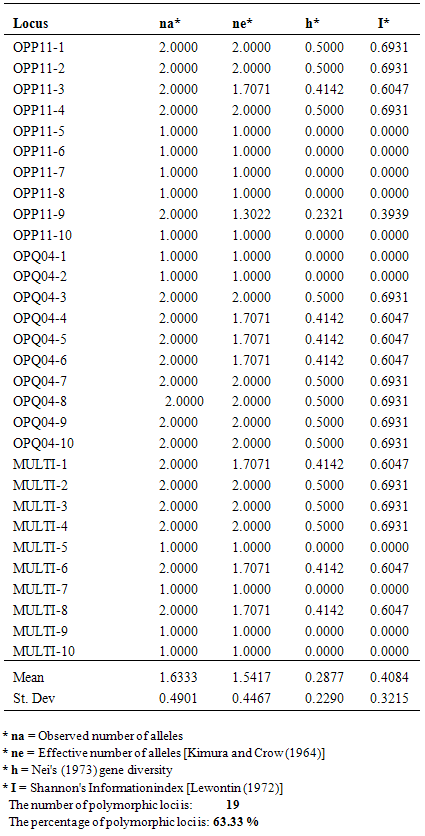
- This study was carried out in six (6) geographically bound states in Nigeria (Nasarawa, Kogi, Benue, FCT, Niger and Plateau State) and the states were assigned population number 1 to 6 respectively. DNA was extracted from fresh young leaves of twenty five (25) cultivars of eggplants randomly sampled across the populations using DNAZOL method following standard protocol (Invitrogen®, 2014). The extracted DNAs were PCR amplified using the following RAPD primers: OPP-11, OPU, B-15, B-18 and OPQ-07. Amplification was done on a thermal cycler (Applied Biosystem in Life Technology version 2720). A total of 75 reactions were carried using customized PCR premix reagent. Each PCR reaction mixture contained 1µl of DNA, 18µl of molecular water and 1µl of one specific primer pair mixed in a PCR tube already containing a customized BIONEER® Accupower PCR Premix reagent to make a total of 20µl as the reaction volume. Temperature gradients were programmed on the thermal cycler following the method of Sifau et al. (2014) and each set of primer reaction lasted for 2 hours. DNA bands were resolved in an agarose gel based electrophoresis tank (Galileo Bioscience tank connected to Consort EV243 electrophoresis power supply) and viewed on a UV based illuminator. Binary matrices manually computed from the bands (using 0and 1 for absence and presence of band respectively) were analysed using POPGEN32 software (1.31 version). Genic Variation Statistics (GVS) for all loci in each population as well as the multi population was analyzed based on their respective number of loci (nl); observed number of alleles (na*); effective number of alleles (ne*); Nei’s gene diversity (h*); Shannon’s diversity index (I*); Number of polymorphic loci and percentage polymorphic loci. Cluster analysis was performed on the nested binary matrices to generate a single dendrogram based on Nei’s (1973, 1987) genetic distance using UPGMA method (Unweighted Pair Group method with Mathematical Average).
3. Results and Discussion
- A DNA band profile of OPP-11 is shown in figure 1. Nei’s gene diversity, as a function of the effective number of alleles in population 1 to 6, that is: 0.288, 0.162, 0.145, 0.007, 0.276 and 0.196 respectively (table 1). Nasarawa State had the highest percentage polymorphism of 63.3% (table 1 and 2) followed by Plateau State (53.3%) and Niger State (43.3%). Kogi and Benue States recorded the same polymorphism of 40% while FCT was the lowest (16.67%). The combined multi population GVS had Nei’s gene diversity of 0.245 and a total number of 25 polymorphic loci which yielded an elevated polymorphism of 83.33%. Though earlier findings on population genic studies on eggplants are lacking, this result is in agreement with the view of population geneticists that effective number of alleles and number of polymorphic loci are major contributory factors that may alter the genotypic frequencies of populations (Taylor et al., 2007). Based on this information, FCT with low polymorphism of 16.67% can be described as a fragile population of eggplant cultivars currently witnessing a phenomenon of genetic drift, therefore unstable. This may be attributed to the political atmosphere and pubic administrative function of the location where farming activities are limited. Therefore, there is need for urgent conservation of the genetically drifted eggplant accessions in this region.
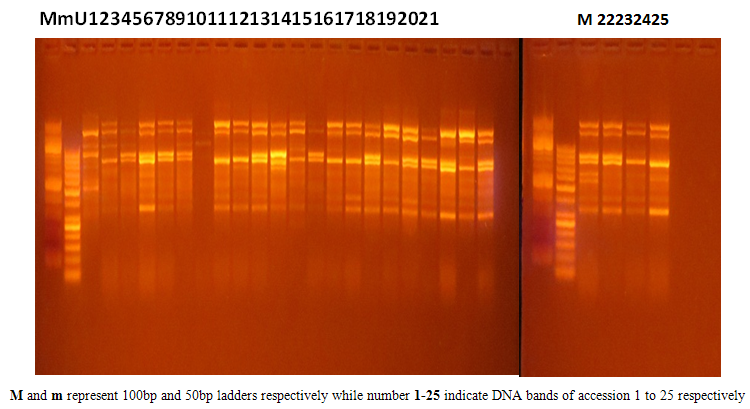 | Figure 1. DNA band profile of OPP-11 |
|
 | Figure 2. Dendrogram of the six populations (six states) based on Nei’s genetic distance |
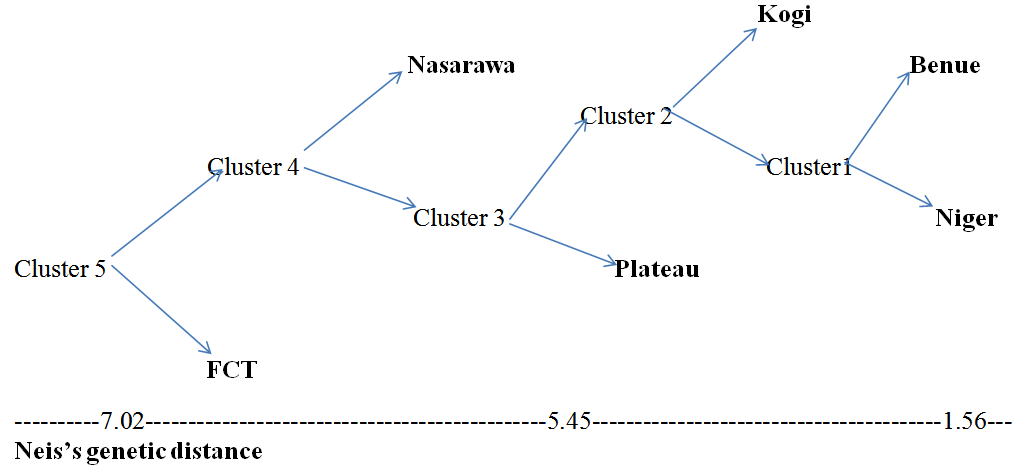 | Figure 3. Tree diagram of genetic relatedness of populations based on Neis’s genetic distance |
|
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML