-
Paper Information
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
International Journal of Plant Research
p-ISSN: 2163-2596 e-ISSN: 2163-260X
2012; 2(4): 131-137
doi:10.5923/j.plant.20120204.05
Genetic Diversity among Some Cucurbits Species Determined by Random Amplified Polymorphic DNA RAPD Marker
Ismail A. Mohammed1, Abdel gabbar N. Gumaa2, Nesreen M. Kamal3, Yasir S. Alnor3, Abdelbagi M. Ali3
1Department of Botany and Agricultural Biotechnology, Faculty of Agriculture, University of Khartoum, Sudan
2Department of Biology, Faculty of Education, University of Khartoum, Khartoum, Sudan
3Agricultural Research Corporation, Wad Medani, Sudan
Correspondence to: Ismail A. Mohammed, Department of Botany and Agricultural Biotechnology, Faculty of Agriculture, University of Khartoum, Sudan.
| Email: |  |
Copyright © 2012 Scientific & Academic Publishing. All Rights Reserved.
RAPD markers were used to determine the genetic relationships and evaluating similarity among some cucurbits species. Thirteen RAPD primers were used to amplify DNA extracted from the leaves of 10 cucurbit species using CTAB method. A total of 227 bands were amplified of which 225 showed polymorphism among the 10 species. PCR-RAPD analysis showed a number of differences in the size and number of bands among the species, which means that there are genetical differences among the studied cucurbit species. Based on these markers, genetic similarity coefficients were calculated and a dendrogram was constructed. The dendrogram analysis delineated three major clusters. The first cluster consisted of one group which comprised Cucurbita moschata and C. pepo at a level of 38.6 % genetic similarity. The second cluster consisted of four groups: Group I comprised Luffa aegyptiaca at a level of 20.6 % genetic similarity. Group II comprised the closely related species Cucumis melo var. reticullatus and C. melo var. flexuosus at a level of 62 % genetic similarity. Group III consisted of Cucumis sativus with about 37.8 % genetic similarity to group II. Group IV consisted of Ctenolepis cerasiformis at a level of 26.6 % genetic similarity. The third cluster consisted of two groups. Group I comprised Citrullus lanatus and Colocynythis vulgaris at a level of 36.2 % genetic similarity. Group II consisted of Coccinia grandis at the level of 19.4 % genetic similarity. The three clusters were similar to each other at a level of 15% genetic similarity. Genetic similarity ranged between 15% and 62 %. This study demonstrates that RAPD markers are useful in assessing genetic diversity among cucurbits.
Keywords: Cucurbits, Genetic Diversity, RAPD Marker
Cite this paper: Ismail A. Mohammed, Abdel gabbar N. Gumaa, Nesreen M. Kamal, Yasir S. Alnor, Abdelbagi M. Ali, Genetic Diversity among Some Cucurbits Species Determined by Random Amplified Polymorphic DNA RAPD Marker, International Journal of Plant Research, Vol. 2 No. 4, 2012, pp. 131-137. doi: 10.5923/j.plant.20120204.05.
1. Introduction
- The family Cucurbitaceae belongs to the order Cucurbitales, class Magnoliopsida (subclass Rosidae). Although most of its species originated in the Old World[1], many originated in the New World, and at least seven genera in both hemispheres[2]. There is tremendous genetic diversity within the family in tropical, subtropical, arid deserts and temperate regions. Archaeological evidence indicated that cucurbits were present in ancient and prehistoric cultures. Molecular markers can be an effective mean to determine genetic relatedness among cultivars and strains used in cucurbits breeding programmes. A number of studies have been designed to examine genetic diversity and phylogenetic relationships among cucurbit cultivars. Some of these studies used biochemical methods such as isoenzyme[3], while others used molecular approaches based on DNA[4] to evaluate genetic diversity in crop species. Most of the isoenzymes tested produced monomorphic patterns ([5,6].Several methods are now available concerning molecular markers studies, and scientists can choose the method of particular interest, depending on available material andobjectives. Among the existing molecular approaches, random amplified polymorphic DNA (RAPD) is one of the most widely used molecular methods in genetic studies and has been applied for the less-known species. However, this method is less reproducible and shows a lower degree of variability as compared with to other methods. RAPD procedure provides a sufficient number of informative markers that could distinguish watermelon cultivars[7, 8]. In a study[9], genetic diversity and relatedness were examined among Citrullus lanatus var. lanatus, C. lanatus var. citroides, and C. colocynthis using RAPD analysis. RAPD markers were also used in the construction of an initial genetic linkage map for watermelon (Citrullus lanatus)[10], and to determine genetic relatedness among Asian watermelon cultivars and breeding lines[11]. Reference[12] used RAPD to study polymorphism and to construct a linkage map for watermelon (Citrullus lanatus (Thunb.) Matsum and Nakai by using 148 primers. Amnon et al.[13] used RAPD markers to study genetic diversity among accessions of some American cultivars of watermelon (Citrullus lanatus var. lanatus). The objectives of this study was studied the genetic diversity among some Cucurbits and the utility of the RAPD markers system for evaluating similarity.
2. Materials and Methods
- Plant material: Accessions of ten cucurbit species were obtained from various sources (Table 1). For DNA extraction the plants were grown in pots.RAPD analysis: For RAPD analysis DNA was extracted from cucurbits leaves according to[14]. Purity and concentration of DNA was tested spectrophotometrically at wave lengths of 260 and 280 nm (Eppendorf – Germany). RAPD reactions were performed using 13 random primers (Nine of these primers were 10-mers (Operon Technologies; OP, and University of British Columbia, UBC), while the other 4 were 12-mers (Bexnet, Co., Japan.) according to[15] using PCR thermocycler. Amplification was performed using touchdown annealing temperature along two or three steps, starting with the melting temperature of the primer and then reducing the temperature by 3 degrees for each step. After all cycles had been completed, 7 µl of the products were mixed with 1 µl of DNA loading buffer and loaded into 2% agarose gel. The agarose gel was then run in 1x TAE buffer at 100 V for one and a half hour. The gels were soaked in 3% ethidium bromide for 30- 60 minutes, and then placed on the UV- transilluminator for band visualization. Cluster analysis: The RAPD fragments 200 to 1500 base pairs (bp) were visually scored as present (1) or absent (0). A dendrogram was constructed based on similarity matrix data by applying the UPGMA[16] and cluster analysis using the software package NTSYS – PC version 2[17].
3. Results and Discussion
- The DNA concentration of the 10 accessions ranged between 80.8 and 352.1 µg/ml, and purity ranged from 1.7 to 2.3 (Table 2). The analysis of the 10 accessions with 13 RAPD primers produced a total of 227 reproducible bands (Table 3). Among these, 225 bands were polymorphic (99.1%) and ranged in size from 200 to 1500 bp. Heikal,et al.[18]reported that the total number of reproducible fragments among the fourteen genotypes of Cucurbita species by six primers reached 463 bands out of them 405 (87.5%) were polymorphic fragments. They found the levels of polymorphism among the fourteen genotypes of Cucurbita species are relatively high (69.8% - 97.0%), indicating the wide gene pool existence in the fourteen genotypes. The total number of bands per primer ranged between 9 and 24, with an average of 17.5 bands per primer (Table 3). The number of polymorphic bands per primer varied from 9 to 24, with an average of 17.3 bands (Table 3). The number of polymorphic bands produced per primer was relatively high among the species (Figs 1 and 2). This high level of RAPD markers polymorphism is in accordance with the results of[19] and[20] who reported that Cucurbita pepo and Cucurbita maxima are highly polymorphic species. However, DNA polymorphism has been reported as moderate to high in other allogamous genera such as Brassica, Allium, Asparagus and Cucumis. Reference[21] found high level of the polymorphism among the different Cucurbita species (97.44%). However, in case of Citrullus lanatus,[22] reported an average of polymorphic bands produced per primer was 21.2.To estimate genetic diversity among the accessions, genetic similarity coefficients were calculated, and a dendrogram was constructed (Fig 3). The range of DNA similarity in the dendrogram was between 0.15 and 0.62. Reference[23] reported a range 0.13 - 0.41 of dissimilarity in 31 landraces of Cucurbita moschata using 31 random primers. The wide range of dissimilarity values suggests that germplasm collection represent a genetically diverse population.The dendrogram consisted of three major clusters. The first cluster consisted of one group (Group І) which comprises the species: Cucurbita moschata and C. pepo which both have a DNA of a genetic similarity of 38.6 %The second cluster consisted of four groups as follows: Group І comprises the species Luffa aegyptiaca which has a genetic similarity of 20.6 %. Group II comprises the closely related species Cucumis melo var. reticullatus and C. melo var. flexuous with a genetic similarity of 62 %. Group III consists of Cucumis sativus. This group is similar to group II in their genetic similarity (37.8 %). Group IV consists of Ctenolepis cerasiformis with a genetic similarity of 26.6 %. The third cluster consisted of two groups. The first group comprises the species Citrullus lanatus and Colocynthis vulgaris which had a genetic similarity of 36.2 %. The second group consists of Coccinia grandis which has a genetic similarity of 19.4 %. Reference[24] analysed accessions of Cucurbita moschata with 11 SRAP primers and reported a total of 148 reproducible bands. Among these, 98(66.2%) were polymorphic and ranged in size from 140 to 950 bp. The total number of bands per primer ranged between 9 and 21, with an average of 13.5 bands per primer. They also analysed accessions of the same plant with 6 AFLP primers and identified a total of 156 reproducible bands which ranged in size from 60 to 380 bp. Of these, 134 (85.9 %) were polymorphic. They found a range of 14 to 41 amplified bands per primer, with an average of 26 bands. Reference[25] analysed accessions of three African edible-seeded cucurbits (Citrullus lanatus L. Matsumra & Nakai, Cucumeropsis mannii L. Naudin and Cucumis melo var. agrestis L. Naudin.), with 11 ISSR primers on DNA extracted from an accession of the three species and observed 66 bands with 4 to 11 bands per primer.Reference[13] analysed accessions of Citrullus lanatus var. lanatus using 138 RAPD primers that were initially screened among some cultivars of watermelon. Of the 138 RAPD primers, only 35 primers produced polymorphic RAPD patterns. Of the later, 25 primers produced 288 reproducible RAPD bands that ranged in size from 100 to 3000 bp.Reference[13] also analysed accessions of Citrullus lanatus and C. colocynthis with 30 RAPD primers that were used for genetic diversity among the two species. The primers produced 662 bands that could be rated with high confidence and ranged in size from 100 to 2650 bp. The number of bands produced by each primer in the above study was relatively high (an average of 22 bands per primer).Reference[26] analysed molecular variation in accessions of Cucumis melo L. and C. metuliferus by 18 RAPD primers and found a total of 176 bands. The number of polymorphic bands was 75 in Cucumis metuliferus and 96 in Cucumis melo. In general, the cluster analysis grouped the accessions according to many morphological traits and showed a clear separation between the species. The RAPD marker revealed considerable genetic diversity among the species, a finding which strongly agrees with the great morphological variability observed among the cucurbits in this study. Similar results were observed for the oriental Cucurbita moschata using RAPD marker[27]. RAPD marker systems proved to be useful for analyzing the genetic diversity of some genera of the family Cucurbitaceae. The knowledge of the diversity of this germplasm will facilitate its use in breeding programmes and the improvement of management of large collections of the species of this family.
|
|
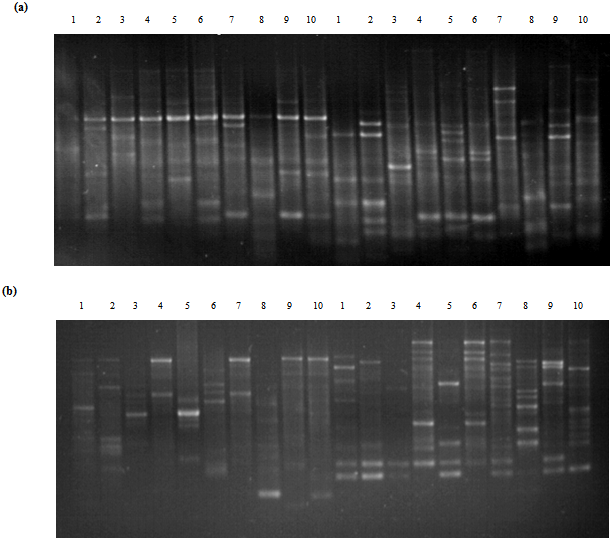 | Figure 1. Agarsoe gel showing polymerase chain reaction (PCR) products of 10 accessions of Cucurbits (a)using UBC157 primer (left) and UBC155 primer (right) and (b) using UBC 222 primer (left) and UBC OPB 05 primer (right) |
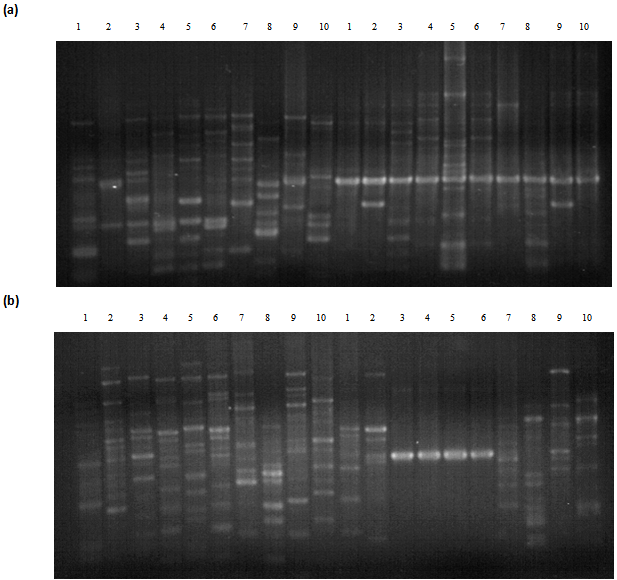 | Figure 2. Agarsoe gel showing polymerase chain reaction (PCR) products of 10 accessions of Cucurbits (a) using A0 2 primer (left) and A01 primer (right) and (b) using A0 4 primer (left) and A0 3 primer (right) |
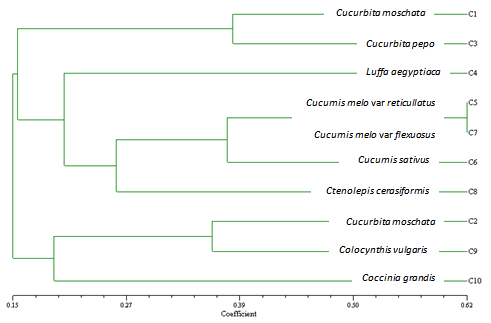 | Figure 3. Dendrogram analysis of 10 species of the family Cucurbitaceae |
References
| [1] | Whitaker , T. W . and G . N. Davis, Cucurbits : Botany , Cultivation and Utilization, 250p,.: Interscience , New York, 1962. |
| [2] | Esquinas-Alcazar, J.T. and. P.J. Gulick, Genetic Resources of Cucurbitaceae, International Board for Plant Genetic Resources, Rome, 1983. |
| [3] | Vallejos, E., Enzyme activity staining. In: S. Tanksley.S.D. and Orton.T.J. (Ed), Isozymes in plant genetics and breeding, Part A : pp. 469-516. Elsevier Science Publisher,1983. |
| [4] | Evett, I.W. and B.S. Weir, Interpreting DNA Evidence. Sinauer Associates Inc.:Sunderland, Massachusetts, USA,1998. |
| [5] | Zamir, D.; Navot, N. and J. Rudich, Enzyme polymorphism in Citrullus lanatus and C. colocynthis in Israel and Sinai, Plant Systematic and Evolution, 146,163-167,1984. |
| [6] | Biles, C.L.; Martyn, R.D. and H.D. Wilson, Isozymes and general proteins from various watermelon cultivars and tissue types, Horticultural Science, 24, 10-812, 1989. |
| [7] | Hashizume, T.; Sato, T. and, M. Hirai, Determination of genetic purity of hybrid seed in watermelon (Citrullus lanatus) and tomato (Lycopersicon esculentum) using Random Amplified Polymorphic DNA (RAPD),Japanese Journal of Breeding, 43:367-375, 1993. |
| [8] | Zhang, X. P.; Rhodes, B.B. and H.S. Skorupska, RAPD Molecular markers in watermelon, Cucurbit Genetics CooperativeReport,17,116-119, 1994. |
| [9] | Levi, A.; Thomas, C.E.; Keinath, A.P. and T.C. Wehner, Estimation of genetic diversity among Citrullus Accessions using RAPD markers, Acta Horticulturae, 510,385-390, 2000. |
| [10] | Hashizume, T.; Skimamoto, I.; Harushima, Y.;Yui, M.; Sato, T.; Imai, T.and M. Hirai, Construction of a linkage Map for watermelon (Citrullus lanatus (Thunb.) Mastsum, & Nakai) using Random Amplified Polymorphic DNA (RAPD), Euphyica, 90, 265-273, 1996. |
| [11] | Lee, S.J.; Shin, J.S.; Park, K.W. and Y.P. Hong, Detection of genetic diversity using RAPD- PCR and sugar analysis in watermelon (Citrullus lanatus (Thunb.) Mansf.) germplasm,Theoretical and Applied Genetic, 92,719-725, 1996. |
| [12] | Toshiharu, H.; Ikuhiro, S.;Yoshiaki, H.; Mamiko, Y.; Takanori, S.; Tsuyoshi, I. and H. Masashi, Construction of a linkage map for watermelon (Citrullus lanatus (Thunb.) Mastsum & Nakai) using Random Amplified Polymorphic DNA (RAPD). Euphyica, 90, 265-273, 1996. |
| [13] | Amnon, L.; Claude, E.; Anthony, P. and T. C. Wehner, Genetic diversity among watermelon (Citrullus lanatus) and Citrullus colocynthis accessions, Genetic Resoures and Crop Evolution, 48,559-566, 2001. |
| [14] | Dellaporta, S.L.; Wood, J. and I. B. Hicks, A plant DNA Mini Preparation. Version 111, Plant Molecular Biology, 18,21-22,1983. |
| [15] | Williams, J.G.K.; Kubelik, A.R.; Liuak, K.J.; Rafalski, J.A. and S.V. Tiney, DNAPolymorphisms amplified by arbitrary primers are useful as genetic markers, Nucleic AcidsResearch,18,6531-6535,1990. |
| [16] | Sneath, P.H.A. and R.R. Sokal, NumericalTaxonomy Freeman, W.H., and Co., San Francisco, U. S. A, 1973. |
| [17] | J. Rohlf, NTSYS-PC, Numerical Taxonomy and Multivariate Analysis System. Version 2, Applied Biostatistical Inc, New York,1993. |
| [18] | Heikal, A. Hadia; Abdel-Razzak, H.S. and E.E. Hafez, Assessment of genetic relationships among and within Cucurbita species using RAPD and ISSR markers, Journal of Applied Sciences Research ,4(5), 515 – 525,2008. |
| [19] | Ferriol, M.; Belen, M.P. and F. Nuez, Genetic diversity of some accessions of Cucurbita maxima from Spain using RAPD and SBAPmarkers,Genetic Resoures and Crop Evolution ,50(3), 227 – 238, 2003. |
| [20] | Ferriol, M.; Pico, B. and F. Nuez, Genetic diversity of a germplasm collection of Cucurbita pepo using RAPD and AFLP markers, Theoretical and Applied Genetic, 107, 271 – 282, 2004. |
| [21] | Athanasios, L.T.; Olga, K.; Anastasia, A., George, N. S., Ekaterini, T. and M. Kouksika-Sotiriou, Description and analysis of genetic diversity among squash accessions, Brazilian Archives of Biology and Technology, 52(2),271- 283, 2009. |
| [22] | Levi, A,; Thomas C.E.; Keinath A.D. and T.C. Whener, Genetic diversity among watermelon (Citrullus lanatus) and Citrullus colocynthisaccessions, Genetic Resoures and Crop Evolution, 48, 556 – 559, 2001. |
| [23] | Gwanama, C.; Labuschang. M.T. and A.M. Botha, Analysis of genetic variation in Cucurbita moschata by random amplified polymorphic DNA (RAPD) markers, Euphytica, 113,19 – 24, 2000. |
| [24] | Marría, F.; Belen, P.; Pascual, F. and N. Fernando, Molecular diversity of a germplasm collection of squash ( Cucurbita moschata) determined by SRAP and AFLP markers, Crop Science, 44,653-664, 2004. |
| [25] | Dje, Y., Tahi, G.C.; Zoro Bi IA.; Malice, M.; Baudoin JP. and P. Bertin, Optimization of ISSR marker for african edible-seeded cucurbitaceae species genetic diversity analysis, African Journal of Biotechnology, 5(2),83-87,2006. |
| [26] | Leah, S.; Kovalski, I.; Huang, R.; Anagnostou, K.; Molly, M. and R. Perl-Treves, Molecular variation in melon ( Cucumis melo L.) as revealed by RFLP and RAPD markers, ScientiaHorticulturae,79,101-111,1999. |
| [27] | Jeon, H.J.; Been, C.G.; Hong, K.H.; Om, Y.H. and B.D. Kim, Identification of cucurbitaceaecultivars by using RAPD markers, Journal of The Korean Society for Horticultural Science, 35,449-456, 1994. |
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML