-
Paper Information
- Next Paper
- Previous Paper
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
International Journal of Plant Research
p-ISSN: 2163-2596 e-ISSN: 2163-260X
2012; 2(3): 80-89
doi: 10.5923/j.plant.20120203.06
Bioinformatics Based Comparative Analysis of Omega-3 Fatty Acids in Desert Plants and Their Role in Stress Resistance and Tolerance
Arjun Sham , Mohammed A. M. Aly
Department of Aridland Agriculture, Faculty of Food and Agriculture, United Arab Emirates University, Al Ain, 17555, UAE
Correspondence to: Mohammed A. M. Aly , Department of Aridland Agriculture, Faculty of Food and Agriculture, United Arab Emirates University, Al Ain, 17555, UAE.
| Email: |  |
Copyright © 2012 Scientific & Academic Publishing. All Rights Reserved.
Numerous studies and clinical investigations have been carried out on the metabolism of poly unsaturated fatty acids (PUFAs) in general and on omega-3 fatty acids in particular over the past two decades. They are involved in a number of stress responses in plants, as illustrated by the genomic studies on Arabidopsis thaliana. Alignment of the sequences and phylogenetic analysis followed by motif elicitation provided ample evidences to show that the stress resistance/tolerance genes have identical sequences which may contribute to the same in all or most of the examined species. In this study we have included a diverse array of plants to find out the involvement of the enzyme omega-3 fatty acid desaturase in omega-3 fatty acid synthesis. Overall results suggest that desaturases play a significant role in stress response in the selected plant species such as date palm and jojoba.
Keywords: Bioinformatics, Date palm, Desaturase, Jojoba, Motifs, Omega-3, Stress
Article Outline
1. Introduction
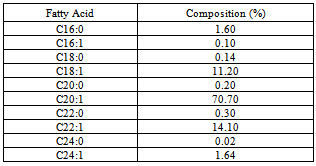
- Over the past two decades, numerous studies and clinical investigations have been carried out on the metabolism of poly unsaturated fatty acids (PUFAs) in general and on omega-3 fatty acids in particular. Omega-3 (ω3) fatty acids are unsaturated essential fatty acids (EFAs). Since these fatty acids are polyunsaturated, the term n-3 PUFAs is applied to them. The term “Omega-3” or n-3 indicates that the first double bond is located at the third carbon from the end of the fatty acid. Omega-3 fatty acids are the bioactive lipids eicosapentaenoic acid (EPA) and/or docosahexaenoic acid (DHA).Jojoba (Simmondsia chinensis) oil is composed almost completely of wax esters of monounsaturated, straight chain acids and alcohols with high molecular weights (C16-C26). Jojoba oil has been defined as a liquid wax ester with the generic formula RCOOR''. RCO represents oleic acid, eicosanoic acid (C20:1), and/or erucic acid (C22:1) moieties. “-OR” represents eicosenyl alcohol (C20:1), docosenyl alcohol (C22:1) and/or tetrasenyl alcohol (C24:1) moieties. Crude jojoba oil contains 0.8 ppm elemental lead (Pb) and less than 0.1 ppm arsenic (AS2S3)[1]. The fatty acid com- position of jojoba oil is presented in Table 1.The date palm tree (Phoenix dactylifera L) besides its known benefits to agriculture over centuries, has been one of the most important staple foods for the native population of Western Persia, Arabia and North Africa. The dates were established within the human nutrition where the studies showed its obvious contribution in the human health as the fruit contains a wide array of fatty acids. Furthermore, the date palm tree has exhibited an outstanding ability to be cultivated in arid areas.
|
1.1. Chemistry of Omega-3 Fatty Acids
- Alpha-linolenic acid (ALA) is the precursor to EPA and DHA. ALA being an 18 carbon omega-3 fatty acid must be converted to 20 and 22 carbon omega-3 fatty acids by enzymes called elongases. In addition, during the conversion of ALA to EPA and DHA more double bonds are added by desaturase enzymes to the molecule during its conversion to the longer chain omega-3. EPA is a 20 carbon fatty acid Which has its first double bond located at the 3rd carbon (counting from the methyl end of the structure). Hence, the location of the first double bond makes EPA an omega-3 fatty acid because the first double bond in its structure end is in the 3rd position. EPA is found in fish and fish oils. Docosahexaenoic acid (DHA) is another omega-3 fatty acid, also found in marine food sources and supplemented in several food products.
1.2. Metabolism of ALA to EPA and DHA
- Figure. 1 illustrates the metabolism of ω-3 fatty acids. For the conversion of ALA to EPA, carbon atoms need to be added to the molecule in the elongation process. Additional double bonds are also added by the desaturase enzymes. ALA has 3 double bonds but EPA has 5 and DHA has 6 double bonds. The process that adds double bonds is desaturation. The enzymes responsible for this process are found in the liver of humans and other mammals (e.g. dogs and cats).A double bond is added at the C6 position of omega-3 alphalinolenic acid by D6desaturase enzyme to form stearidonic acid. And, the chain is elongated by the addition of 2C units by elongase enzyme until a 20 carbon atom containing compound is formed. This fatty acid undergoes desaturation at C5 by D5desaturase forming the omega 3 fatty acid. By alternate desaturation and elongation by desaturaes and elongases respectively a 24 carbon containing fatty acid is formed. This, on oxidation, forms docosahexaenoic acid which is an omega-3 fatty acid.
1.3. Nutritional Significance of Date Palm
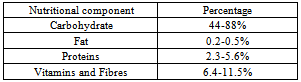
- The flesh of date fruit contains 0.2-0.5% oil, whereas the seed contains 7.7-9.7% oil (Table 2.). The weight of the seed is 5.6-14.2% of the date. Unsaturated fatty acids in date palm include palmitoleic, oleic, linoleic and linolenic acids. The oleic acid content of the seeds varies from 41.1 to 58.8%, which suggests that the seeds could be used as a source of oleic acid.
1.4. Types of Stress in Desert Plants
1.4.1. Temperature Stress
- Temperature stresses experienced by plants can be classified into three types: (a) temperatures below freezing, (b) low temperatures above freezing, and (c) high temperatures. Each plant species has its own optimum temperature for growth, and its geographical distribution is determined to a major extent by the temperature zone in which it can survive[2]. Rapid advances in molecular genetics have allowed gene cloning from both prokaryotes and eukaryote, which provided understandings of the mechanism of temperature adaptation[3, 4]. When plants are exposed to salt, drought, and low-temperature stresses, they accumulate highly soluble organic compounds of low molecular weights, called compatible solutes. Typical compatible solutes include mannitol and other sugar alcohols, amino acids such as proline, and amino acid derivatives such as glycinebetaine. Some of these compatible solutes, e.g. proline, are accumulated practically in all plant species, whereas others like glycine and betaine are distributed only among plants with a high tolerance to salt or cold temperatures[5].
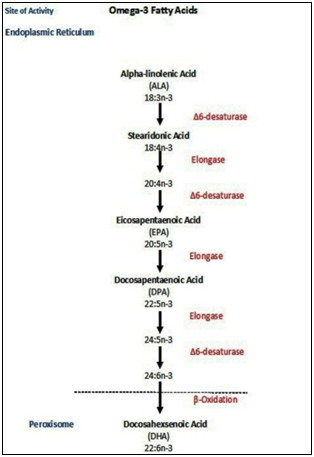 | Figure 1. Metabolism of omega-3 fatty acids |
|
1.4.2. UV Stress
- Sunlight contains energetic short wavelength ultraviolet (UV) photons which are potentially detrimental because of their destructive interactions with many cellular molecules, such as the amino acids of essential proteins, nucleic acids bases or membrane lipids[10]. The destructive action of UV irradiation is the result of both direct and indirect mechanisms involving endogenous sensitizers and the generation of active oxygen species. Physiological and biochemical effects of UV-B radiation include effects on enzymes, stomatal resistance, concentrations of chlorophyll, protein and lipid, reduction in leaf area, and tissue damages[11,12]. Some plants, however, appear to be quite resistant to increased UV irradiation. The differential susceptibility of plants to UV stress is clearly an important factor in their competitive relationships in terrestrial ecosystem.
1.4.3. Drought Stress
- Desert plants generally follow two main drought stress adaptation strategies. They tolerate the drought through phenologic and physiological adjustments referred to as tolerance or avoidance of drought through dormancy[13]. Both tolerance and avoidance mechanisms contribute to the ability of a plant to survive drought, but it also depends on the frequency and severity of the drought periods[14, 15]. Plants under such stress conditions regulate their water status using several tactics viz., osmotic adjustment, stomatal aperture, turgor maintenance, root distribution and leaf canopy properties[16]. Leaves developed under drought conditions generally exhibit small cell size, thick cell walls, small vacuoles and higher concentration of osmotica[17]. The water relation parameters, therefore, do not differ significantly between young and old leaves[18]. Plant tolerance to high temperatures and drought stresses is primarily achieved by preventing water loss through decreasing internal water potential and closing stomates. The published literature reveals that molecular and physiological basis of drought stress tolerance in organisms is very well researched[19, 20, 21, 22, 23, 24, 25, 26, 27, and 28]. In general, response to protect cells from dehydration stress at molecular level, stress induced proteins or amino acids (e.g. late embryogenesis abundant (Lea) proteins, heat shock proteins (Hsp), proline, glycine betain), osmoprotectant molecules (e.g. sugars like trehalose, mannitol, glycerols), enzymes that improve membrane flexibility like omega 3 fatty acid desaturase, and scavengers of reactive intermediates superoxide dismutases, peroxidases, glutathion reductases, catalases are involved. Specific responses vary with the organisms.
1.4.4. Salinity Stress
- Height salt concentration normally impair the cellular electron transport within the different subcellular compartments and lead to the generation of Reactive Oxygen Species (ROS) such as singlet oxygen, superoxide, hydrogen peroxide and hydroxyl radicals[29, 30, 31]. Excess of ROS triggers phytotoxic reactions such as lipid peroxidation, protein degradation and DNA mutation[32, 33, and 34]. Mannitol is accumulated by a wide range of species in response to salinity[35].The ectopic expression of the mtlD gene for the biosynthesis of mannitol improves tolerance to water stress and salinity. The Salt-Overly-Sensitive (SOS) pathway that connects a calcium signal through a calcium-binding protein (SOS3) via a kinase transmitter (SOS2) to a sodium exclusion (SOS1) function[23] has been established as a major ionic (Na+) stress regulatory system. There is currently no evidence for a relation between the co expression of mannitol and omega-3 although both are involved in salinity stress response.
1.4.5. Reactive Oxygen Species (ROS)
- When different pathways are uncoupled, electrons that have a high-energy state are transferred to molecular oxygen (O2) to form reactive O2 species (ROS);[36, 37]. It is known that the oxidative stress response is triggered by an imbalance in the production and metabolism of ROS. A variety of stress conditions, both biotic and abiotic, trigger a transient, enzyme-mediated increase in ROS, predominantly O2 – and H2O2, in plant and animal cells. The transient output of ROS was termed oxidative burst. Temperature stress such as heat, cold or freezing is a principal cause for yield reduction in crops[38]. ROS generated by these stresses have been shown to injure cell membranes and proteins[39, 40]. An additional contributor to cellular damage during temperature stress is high light. High-light stress has the potential to enhance the production of ROS in cells and cause oxidative damage to chloroplasts[41].
1.5. Omega-3 Fatty Acids and Stress
- The analyses of Arabidopsis mutant strains have shown that polyunsaturated fatty acids in chloroplast membranes influence the size of the chloroplast and the formation of its membranes at low temperatures[42]. The fad5 mutant is deficient in the activity of a chloroplast omega-9 fatty acid desaturase and accumulates high levels of palmitic acid (16:0). Conversely, the fad6 mutant is deficient in the activity of the chloroplast omega-6 fatty acid desaturase and accumulates high levels of 16:1 and 18:1 fatty acids. Both mutants show correspondingly reduced levels of polyunsaturated fatty acids in the chloroplast galactolipids. These observations suggest that polyunsaturated fatty acids are required for surviving low temperatures.The jojoba requires a minimum of established techniques and maintenance in the first stages of its proliferation and it has deep root systems and grows in both arid and saline conditions[43, 44]. The tolerance of jojoba to abiotic stresses has been studied[45]. Salinity was reported to cause reduction of the elongation and the thickening of stems[46, 47], reduction of the shoot system and leaf size as well as an increase in the thickening of leaves[45]. Salinity also influenced cutinisation, reduced the development of vascular tissues and increased the density of the trichomes[48] and the chemical composition of fatty acids and fatty alcohols in jojoba seeds[49].Kayani et al.[49] examined jojoba seeds germinated in an artificially salinized sand medium in a greenhouse at 32°C. Seeds and seedlings were sampled for fatty acid and fatty alcohol contents at 3, 6, 10, 15, 20, 25 and 30 days after sowing. The relative proportions of palmitic acid, eicosenoic acid, tetracosenoic acid and tetracosenol among the treatments were found not to differ significantly. Increasing salt concentration was found to result in decreased amounts of oleyl alcohol, eicosenol and oleic acid and increased amounts of docosenol, hexacosenol, docosenoic acid and stearic acid. These changes were not observed in the first 20 days and were noted only in the last 10 days of the experiment. They concluded that the salinity of the germination medium induced some changes in the composition of lipids in jojoba seeds and that the changes were probably the result of an indirect effect of salinity on lipid utilization.With the available information on A. thaliana stress responsive genes, an attempt was made to find out if the desaturase gene present in jojoba has any effect on the stress response. The sequence of the desaturase gene in both Arabidopsis (A. thaliana) and jojoba (Simmondsia chinensis) were collected and performed a similarity search using blastn. By obtaining positive results on blasting the sequences, we performed a motif analysis using insilico methods to find out the presence of common motifs in the fatty acid gene coding sequences. Motifs indicate regular repeating units which are present in different species. In the present study, motifs identification and analysis provided information on the stress related genes which encode for omega-3 fatty acid biosynthesis.
2. Materials and Methods
2.1. Using BLASTn
- All the sequences were aligned using the BLASTn, which is a sub-division of the online tool BLAST (Basic Local Alignment Search Tool). This was obtained from www.ncbi.nlm.nih.gov/BLAST[50] illustrating the score value, total score, and percentage of query coverage and E-value of the alignment. The blastn results examined the query sequence of Fatty Acid Desaturase from Arabidopsis thaliana against non redundant databases of Simmondsia chinesis, Zea mays, Arabidopsis thaliana, Glycine max and Phoenix dactylifera[50].
2.2. Multiple Sequence Alignment
- The alignment was confirmed using ClustalX2 by introducing gaps. The Clustal series of programs are widely used for multiple alignments and for preparing phylogenetic trees. Clustal tries to align the most-closely related sequences first[51], in order to build a representative profile of the family. Clustal X can calculate trees by using the Neighbour-Joining method[52], a widely used and relatively fast algorithm that clusters sequences by minimizing the sum of branch lengths.
2.3. Phylogenetic Analysis
- NJplot is able to draw any phylogenetic tree expressed in the Newick phylogenetic tree format. NJplot is especially convenient for rooting the unrooted trees obtained from parsimony, distance or maximum likelihood tree-building methods.
2.4. Motif Analysis
- The presences of motifs in the selected sequences were identified using Multiple Em for Motif Elicitation (MEME) Suite[53]. A motif is a sequence pattern that occurs repeatedly in a group of related protein or DNA sequences. MEME represents motifs as position-dependent letter-probability matrices which describe the probability of each possible letter at each position in the pattern. Individual MEME motifs do not contain gaps. Patterns with variable-length gaps are split by MEME into two or more separate motifs.
2.4.1. Identification of Overlapping Motifs
- MCAST searches a sequence database for statistically significant clusters of non-overlapping "hits" to the motifs in a query. MCAST searches for all of the matches between the query and the sequences in the database. Each match is assigned an E-value, and matches that score below an E-value threshold are printed in order of increasing E-value. The motifs predicted by MEME were compared against Transfac database. The output of TOMTOM is a web page displaying a table of motifs in the database similar to the motif submitted. Each entry in the table includes the database identifier for matching motif, its description, the p-value and q-value of the match, the overlap and offset between your motif and the matching motif, the strand of the matching motif, and a logogram of the motif and the matching motif. The motifs showing similarity to the transfac database sequences are indicated by large alphabets in the output image.
3. Results
3.1. Blast Output
- The output of Blast showed the results of the query sequence of Fatty Acid Desaturase from Arabidopsis thaliana against non redundant databases of Simmondsia chinesis, Zea mays, Arabidopsis thaliana, Glycine max and Phoenix dactylifera (data not shown).
3.2. Clustal X2 Output
- The output of CLUSTALX shows the conserved regions with the gapped alignment of the selected sequences. Asterisks indicate residues exactly conserved across all the sequences, while dots indicate regions where all pairs of residue have a similarity score greater than or equal to 10[51].
3.3. Translation
- The protein sequences of all species other than Phoenix dactylifera were taken from NCBI. The DNA sequences whose translation product are not provided in NCBI were converted to their corresponding protein sequences using the translate tool from ExPasy Proteomics server. The product of translation from the desaturase genes of Phoenix dactiliera (virtual proteins) is obtained by using Translate Tool from ExPasy Proteomics Server[54].
3.4. NJ Tree
- A phylogenetic tree, also known as a phylogeny, is a diagram that depicts the lines of evolutionary descent of different species, organisms, or genes from a common ancestor. Phylogenies are useful for organizing knowledge of biological diversity, for structuring classifications, and for providing insight into events that occurred during evolution. Furthermore, because these trees show descent from a common ancestor, and because much of the strongest evidence for evolution comes in the form of common ancestry, one must understand phylogenies in order to fully appreciate the overwhelming evidence supporting the theory of evolution.The tree diagram (Figure. 2) shows the evolution of different desaturase genes with the divergence occurring from two divisions. In first division Arabidopsis thaliana desaturase (gi|42564197), Simmondsia chinesis desaturase (gi|169894|) evolves independently from one node. The desaturase genes of Oryza sativa form another clade (gi|88606638|). Zea Mays desaturase (gi|226498699) evolved alone from Oryza sativa. Glycine Max desaturase (gi|15430569|) evolved independently from Glycine max omega-3 cultivar (gi|239775531) with Glycine max as a node. The desaturases from P. Dactylifera (PDK_20s1824371g001, PDK_20s1546341g002 and PDK_20s1416541g002) evolved almost simultaneously. Arabidopsis thaliana FAD8 (gi|145357671) and FAD7 (gi|30681624) have evolved simultaneously and recently.
3.5. Motif Prediction Tool- MEME Output
- A motif is a sequence pattern that occurs repeatedly in a group of related protein or DNA sequences. Conserved protein sequence motifs are short stretches of amino acid sequence patterns that potentially encode the function of proteins. The analysis of our sequences using MEME provided 3 different motifs. The output (Figure. 3) shows the summary of motifs predicted by MEME. With the shortest sequence length of 489, longest sequence length of 5326 and average sequence length of 1663.9 residues[53]. The predicted motifs in the sequences are listed in Figure 4. The output of MAST (Figure. 4) showing the region of conserved motifs from the start region, strand in which the motif is present along with the p-value is given below for each of the three predicted motifs[53]..
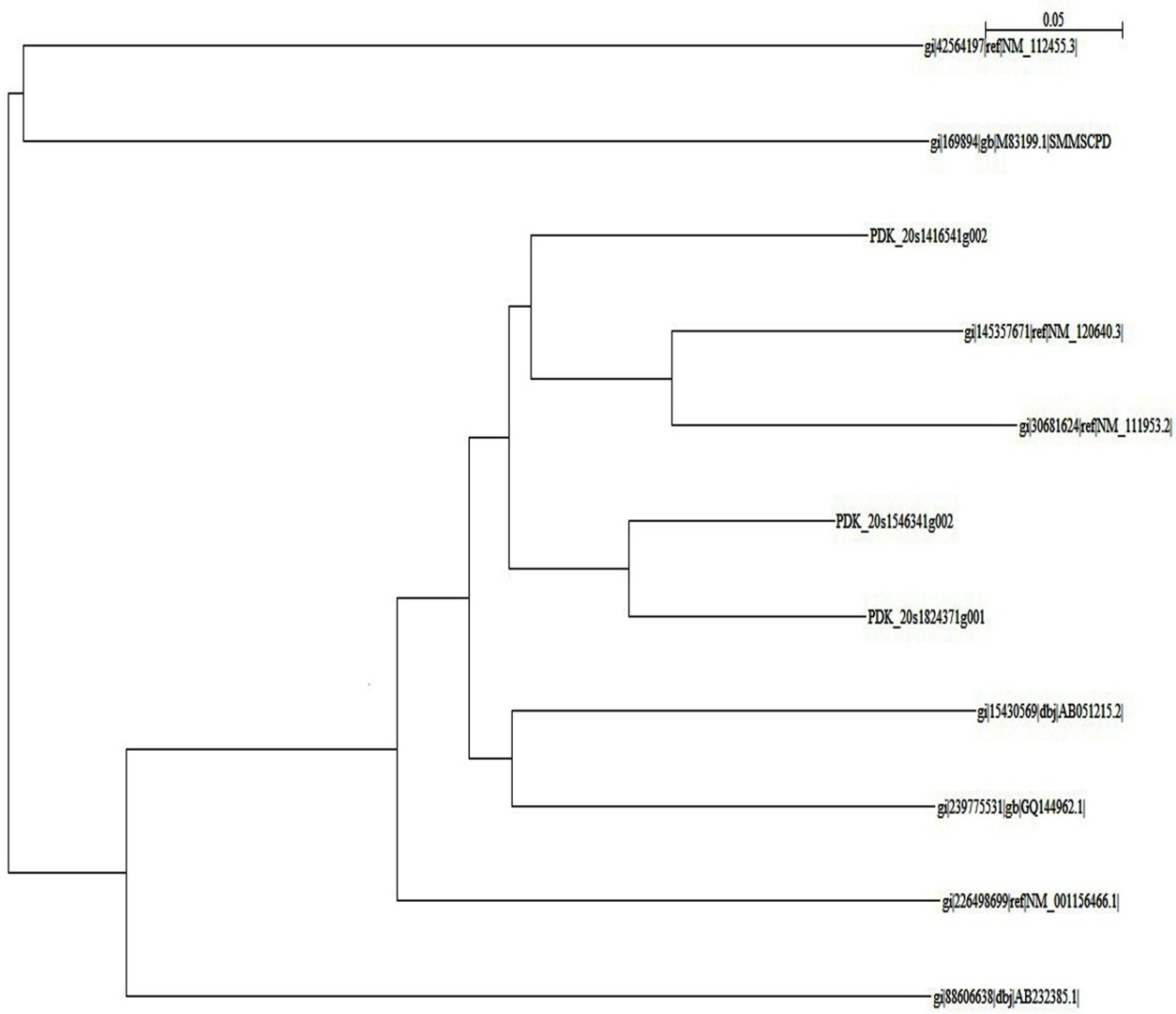 | Figure 2. NJPlot output |
3.5.1. Motif Cluster Alignment and Search Tool (MCAST)
- MCAST searches a sequence database for statistically significant clusters of non-overlapping "hits" to the motifs in a query. MCAST searches for all of the matches between the query and the sequences in the database. Each match is assigned an E-value, and matches that score below an E-value threshold are printed in order of increasing E-value.
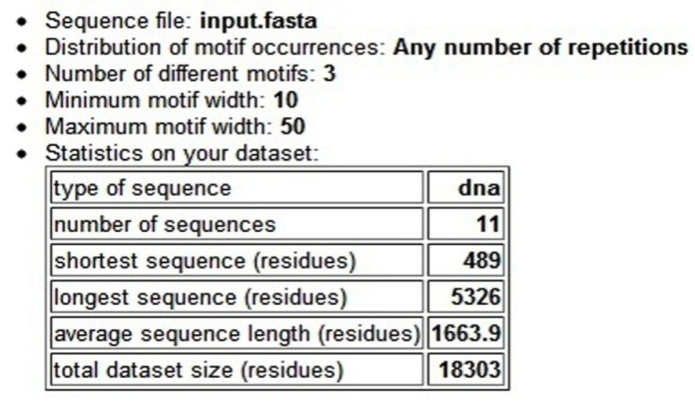 | Figure 3. MEME output summary |
 | Figure 4. MAST output |
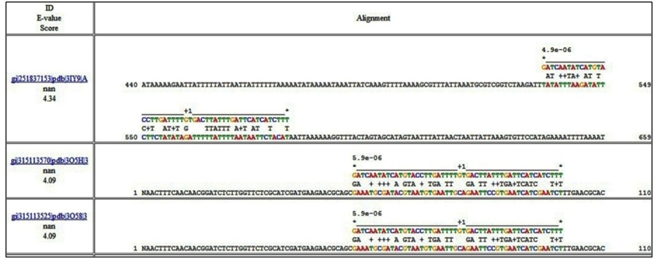 | Figure 5. MCAST alignment results |
3.5.2. Alignment
- Each alignment lists the sequence identifier, match E -value and log-odds score along the left. On the right, it shows the alignment of the match with the sequence in groups of four segments. The bottom-most line in each segment contains 50 letters from the target database sequence, flanked by that segment's start and end locations within the entire sequence. Thus, the first segment would be flanked by "1" and "49", the second by "50" and "99", etc. Aligned above these 50-letter segments are the motifs corresponding to the hits in the match. The motifs are labelled with numbers in the order they appear in the query. A plus or minus sign preceding a hit indicates that the hit occurs on the given (+) or reverse complement (-) of the DNA sequence in the database. Each position within a motif region is indicated by a letter, and each gap position is indicated with a period. In between the sequence segment and the corresponding match positions is a line that indicates the degree of match between the motifs and the sequence. If the letter in the motif with the largest log-odds score appears in the sequence, then the match row contains that letter. If the sequence letter does not have the largest log-odds score, but does have a positive log-odds score, then the match row contains a plus sign. Otherwise, the match row is empty. The top-most row of each segment shows the p-value (or log-odds score) of each hit aligned above the start of the hit, depending on the score mode. MCAST Alignment (Figure. 5) shows the alignment of motifs predicted by MEME[55].
3.5.3. Motif Diagrams
- The motif diagrams section shows the matches in schematic format (Figure. 6). For each match, in the right two columns, it shows the sequence identifier and the match E-value. On the left, it shows the positions and spacing of the hits making up the match. Hits are labelled with numbers corresponding to the order the motifs were given in the query. A plus or minus sign preceding a hit indicates that the hit occurs on the given (+) or reverse complement (-) of the DNA sequence in the database. Figure 6 represent the position of motifs in the DNA sequences from MCAST.
3.6. TOMTOM Output
- The TOMTOM web application compares an input DNA motif to the elements of a database of known motifs (and their DNA reverse complements). A list of matching motifs is reported, ranked by q-value. The q-value is the minimal false discovery rate at which the observed similarity would be deemed significant. The output contains results for each query, in the order that the queries appear in the input file. With respect to each query, targets are ranked by q-value[56].The output (Figure. 7) shows the output of TOMTOM tool of MEME suite. TOMTOM matches the predicted motifs against Transfac matrix. The target sequence, p-value, q-value and E-value are also summarized[56]. The P value is a probability, with a value ranging from zero to one. The ‘e’ value gives a measure of the similarity of sequences, the lower the e value, the higher the congruity of your query sequence and the retrieved sequence. The q-value is the minimal false discovery rate at which the observed similarity would be deemed significant. The output contains results for each query, in the order that the queries appear in the input file. With respect to each query, targets are ranked by q-value.
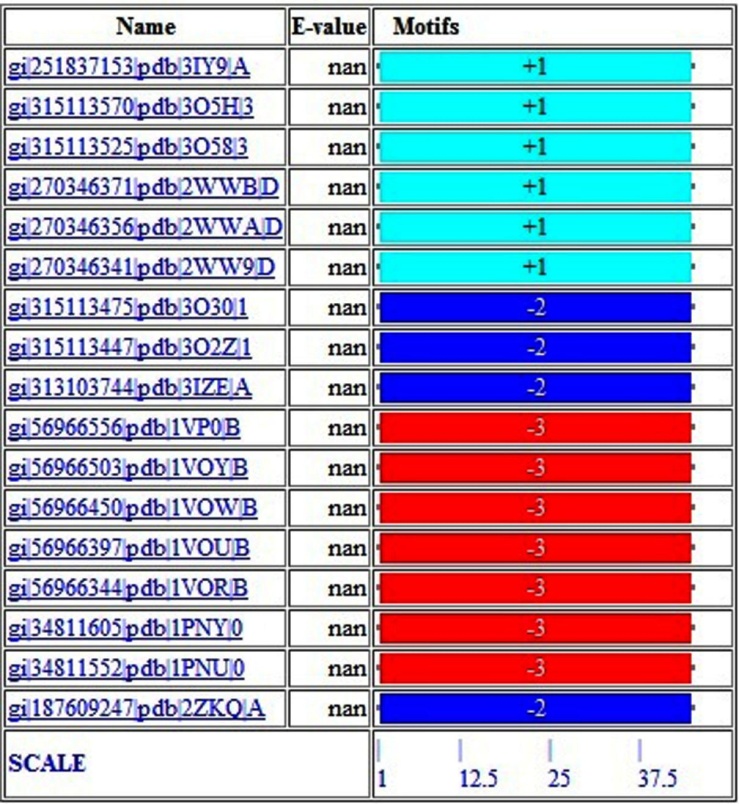 | Figure 6. MCAST motif diagram |
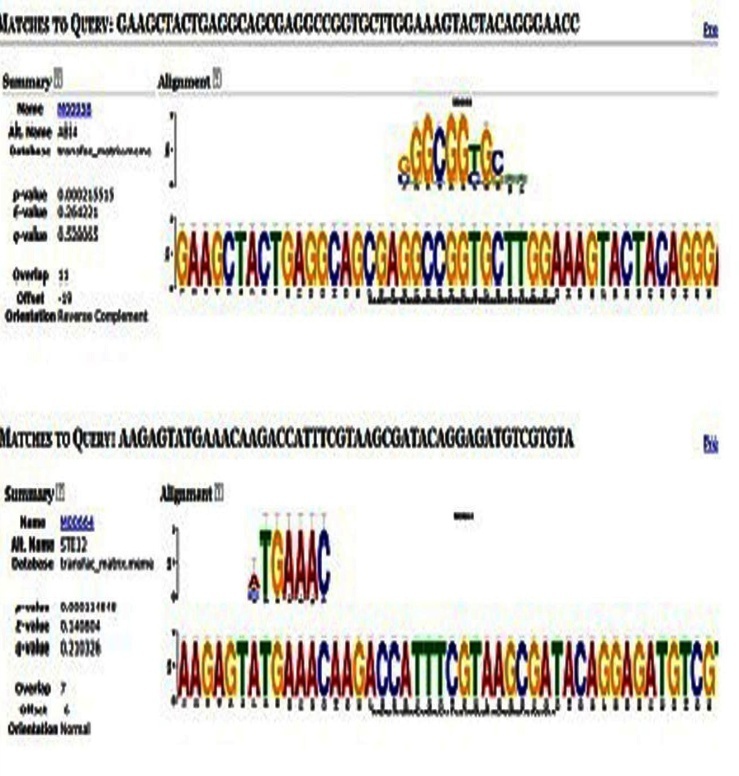 | Figure 7. TOMTOM Output. |
4. Discussion
- In this study we have included a diverse array of plants to find out the involvement of the enzyme omega-3 fatty acid desaturase in omega-3 fatty acid synthesis. Blast results have provided similar sequences related to the query sequences of desaturase we have selected for the study. The multiple sequence alignment showed the conserved regions in all the sequences and based on this a phylogenetic tree was drawn to find out the evolution of these genes (Figure. 2). Since motifs play an important role in the function, the presence of similar motifs in all the sequences indicates a similarity in their response to stress. The sequence similarity in the blast results indicated that the genes coding for omega 3 fatty acid metabolism are similar. Motif analysis indicated that repeated motifs are present in all the selected species in our present investigation. Overall results suggest that desaturases in the examined plant species, e.g. date palm and jojoba play a significant role in stress response.Stress adapted plants respond to abiotic and biotic stress by remodeling membrane fluidity and by releasing α-linolenic (ALA) from membrane lipids. The modification of membrane fluidity is mediated by changes in unsaturated fatty acid levels, a function provided in part by the regulated activity of fatty acid desaturases. Adjustment of membrane fluidity maintains an environment suitable for the function of critical integral proteins during stress. The modulation of chloroplast oleic acid (18:1) levels is central to the normal expression of defense responses to pathogens in Arabidopsis[57]. Oleic (18:1) and linolenic (18:2) acid levels, in part, regulate development and seed colonization[57].Omega-6 and omega-9 are two other fatty acids present in plants. All omega-3, 6 and 9 fatty acids are synthesized via desaturases. All desaturase enzymes have a unique property of being involved in stress resistance/tolerance. Since desert plants such as date palm an jojoba are constantly exposed to a variety of stresses, it is worth noting that omega-6 and omega-9 also play an important role in stress response.Most approaches to molecular breeding have attempted to improve host tolerance to temperature stress by introducing a single gene, thereby altering only a single trait. However, the general view is that multiple traits are involved in tolerance over a wide range of temperatures. Consequently, approaches that aim to simultaneously alter multiple related traits are also being attempted in order to engineer better adaptation to high temperatures[2]. The development of a more effective approach to creating plants that can resist/tolerate a wide range of stresses remains a challenge for the future. To address this issue, it is necessary to identify the key strategies that plants use to deal with complex stresses of both biotic and abiotic origin. With the swift development in recent years of functional genomics, the comprehensive systematic analysis of transcriptomics and proteomics, it will facilitate the discovery of intrinsic resistance/tolerance factors that are vital in allowing plants to cope with a wide range of stresses, including biological as well as physical and chemical environmental stresses. Our investigation supports the possible involvement of omega-3 fatty acids in stress tolerance in desert palnts with reference to date palm and jojoba.
5. Conclusions
- The literature survey reveals the stress tolerance property of desaturase enzyme involved in the synthesis of omega-3 fatty acids. In this study an attempt was made to identify any stress resistance/tolerance property of omega-3 fatty acids in jojoba and date palm based on the concept that similarity in sequence leads to similarity in function. Blasting the sequences of desaturase enzyme of the model organism A. Thaliana with that of desaturase from jojoba revealed that their similarity. Also, a phylogenetic tree was constructed to test the evolutionary conservation of the gene sequence. Motif identification was performed using MEME suite which returned three common motifs in all the sequences both in positive and negative strands. These significant results indicate that the gene encoding desaturase enzyme in jojoba might play a major role in stress resistance.
ACKNOWLEDGEMENTS
- The authors are grateful to the United Arab Emirates University for making this manuscript possible through the NRF research grant No. RSA - 1108 – 00479 (UAE University No. 31F002).
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-Text HTML
Full-Text HTML