-
Paper Information
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
International Journal of Plant Research
p-ISSN: 2163-2596 e-ISSN: 2163-260X
2011; 1(1): 16-24
doi:10.5923/j.plant.20110101.03
Molecular Analysis of a Copper- and Zinc-Containing Superoxide Dismutase Gene Isolated From the Latex of Euphorbia Characias: Another Piece in the Molecular Puzzle of Euphorbiaceae Latex Proteins
E. Atzori1, A. Rescigno2, A. Padiglia1
1Department of Life and Enviroment Sciences, University of Cagliari, Cittadella Universitaria di Monserrato, 09042, Monserrato, Cagliari, Italy
2Department of Biomedical Sciences and Technologies, University of Cagliari, Cittadella Universitaria di Monserrato, 09042, Monserrato, Cagliari, Italy
Correspondence to: A. Padiglia, Department of Life and Enviroment Sciences, University of Cagliari, Cittadella Universitaria di Monserrato, 09042, Monserrato, Cagliari, Italy.
| Email: |  |
Copyright © 2012 Scientific & Academic Publishing. All Rights Reserved.
A copper- and zinc-containing superoxide dismutase (Cu/Zn-SOD) cDNA was isolated from the Euphorbia characias latex (Elx) using consensus degenerate hybrid oligonucleotide primer (CODEHOP) design and RT-PCR strategy. Both 3’and 5’untraslated regions (UTR) were isolated by rapid amplification of cDNA ends (RACE) method. Analysis of the nucleotide sequence revealed that the Cu/Zn-SOD cDNA contains an open reading frame encoding a protein of 152 amino acids. Bioinformatic analyses of Elx SOD gene revealed a high identity rate with a large number of plant Cu/Zn-SODs in the deduced amino acid sequence. Since isoenzymes may be generated through the multiplicity of SOD genes or result from post-trascriptional events, genomic Southern blot in conjunction with northern blot experiments were also performed. The genomic analysis showed that the E. characias genome contains a single SOD gene. Northern blot analyses confirmed the presence of a single SOD mRNA demonstrating that alternative splicing does not occur. Quantitative real time-PCR (qRT-PCR) experiments showed that SOD gene expression in latex reaches maximum levels during the summer. These results suggest that the Cu/Zn-SOD of Euphorbia characias latex probably may be involved in the antioxidative process triggered by oxidative stress induced by the conditions of environmental change in which the plant lives.
Keywords: Euphorbia Characias, Latex, CODEHOP, Cu/Zn-SOD, EF176601, Plant Defense
Cite this paper: E. Atzori, A. Rescigno, A. Padiglia, Molecular Analysis of a Copper- and Zinc-Containing Superoxide Dismutase Gene Isolated From the Latex of Euphorbia Characias: Another Piece in the Molecular Puzzle of Euphorbiaceae Latex Proteins, International Journal of Plant Research, Vol. 1 No. 1, 2011, pp. 16-24. doi: 10.5923/j.plant.20110101.03.
Article Outline
1. Introduction
- Euphorbia characias is a perennial shrub belonging to the Euphorbiaceae family, whose habitat is represented by sunny locations with dry soils characterized by high salinity. Characteristic of all Euphorbiaceae is the presence of laticifers[1,2], specialized secretory cells forming vessel–like structures containing latex, a milky sap with a complex composition. Among the components are many specialized metabolites such as cardenolides, alkaloids, and natural rubber as well as a number of mechanisms by repelling and killing phytopathogens and sealing wounded areas[3,4,5]. Previous studies have confirmed the presence of several enzymatic activities in the Euphorbia characias latex (Elx), and the most relevant enzymes have been purified and characterized. One of these is a cationic peroxidase named ELP (Euphorbia latex peroxidase), a member of class III secreted plant peroxidases[6,7]. These enzymes can oxidize a variety of aromatic molecules such as hydroxycinnamic acid, hydroxycinnamyl alcohol and their derivatives, in the presence of hydrogen peroxide (H2O2)[8]. The H2O2 then participates directly in the plant’s defense responses mediated by the oxidative burst, and is also believed to act as a second messenger for the induction of plant defensive genes[9,10]. Intriguingly, ELP activity was recently shown to be regulated by its interaction with calcium/calmodulin (ELCaM) complexes, the first example of a CaM-binding peroxidase[7]. Moreover, earlier findings suggest that catalase (ECat) and antiquitin (EAq) might represent additional nodes in the Euphorbia defense systems, and a multi-enzymatic interaction contributing to the plant’s protection against biotic and abiotic stresses was proposed to occur in E. characias laticifers[11]. Among antioxidant enzymes, superoxide dismutase (SOD; EC 1.15.1.1) is considered to be a key enzyme in the natural defense against free radicals[12]. SOD mainly removes highly toxic superoxide anion (O2–-) by dismutation into H2O2 and molecular oxygen (O2)[13,14]. According to the structure and metal ions present in the active site centre, three different classes of superoxide dismutases can be distinguished: Cu/Zn-SODs, manganese-containing SOD (Mn-SOD), and iron-contai- ning SOD (Fe-SOD). Moreover, a new superoxide dismutase containing nickel (Ni-SOD) was found in Streptomyces species and cyanobacteria[15,16]. Cu/Zn-SOD is usually present in eukaryotic cytosol, in the outer mitochondrial space and in chloroplasts[17-20];. Mn-SOD is located in prokaryotic cytosol and in the mitochondria of eukaryotes, while Fe-SOD has been found in eubacteria and archaebacteria[21]. The large majority of these SODs require specifically Fe or Mn to perform their enzymatic activity, but only some, like as the themostable cambialistic-SOD cloned and characterized from Antrodia camphorate, function with both types of ions[22]. The Mn-SOD and Fe-SOD, very similar in structure characterized by N-terminal α-helices and a C-terminal α/β domain, seem to have evolved from a common ancestral gene[23,24]. These proteins in general assemble into dimers or tetramers of identical subunits, and crystal structures are now available for several of these enzymes[25-28]. The structure of Ni-SOD reveals a homohexameric structure of four-helix- bundle subunits[29]. The Cu/Zn-SODs are not structurally related to the latter, and therefore some authors consider that these enzymes may represent a different type of SOD. All eukaryotic Cu/Zn-SODs characterized to date are homodimeric enzymes containing one copper and one zinc ion per subunit[30,31]. Several structural studies have shown that these eukaryotic enzymes display a remarkably conserved three-dimensional fold, based on a flattened Greek-Key, eight-stranded β-barrel[32-36,23]. Beyond the structural differences, all SODs share the common role as main controller of cellular reactive oxygen species (ROS) levels. In plants, the role of SOD during environmental adversity has received great interest since O2–- has been found to be produced under various types of stress conditions[37,38]. So far, several molecular studies have been conducted about SODs expressions from various plant families, but not for any Euphorbiaceae. In this paper, we demonstrate the presence of Cu/Zn-SOD in Elx starting from the detection and sequencing of the relevant encoding cDNA. Gene expression was assayed in latex during the seasons by qRT-PCR. The potential interaction of this protein with the other enzymatic systems previously described in Elx is discussed. To our knowledge, this is the first report showing evidence of a Cu/Zn-SOD in Euphorbiaceae.
2. Methods
2.1. Plant Material
- Elx was collected in the four seasons at several locations in southern Sardinia (Italy), frozen, and stored at –80℃ until use. All chemicals were obtained as pure commercial products and used without further purification.
2.2. Isolation of RNA from Latex and cDNA Synthesis
- E. characias branches were sliced and the fresh latex (≈ 5mL). flowed directly into a tube containing 20 mL of 2 RNA extraction buffer (0.1 M Tris-HCl, 0.3 M LiCl, 0.01 M EDTA, 10% SDS, pH 9.5) with 5 mL of RNAlater solution (Sigma-Aldrich, St. Louis, MO) to stabilize and protect RNA. The latex solution was mixed and centrifuged at 8000 g for 15 min at 20 ℃. The supernatant fraction was processed using TRI Reagent RNA isolation reagent (Sigma-Aldrich) according to the manufacturer’s instructions. The quality of purified RNA was verified by gel electrophoresis using 1% denaturing agarose gel stained with ethidium bromide (Sigma-Aldrich) and recording the absorbance spectrum from 220 to 300 nm. To obtain cDNAs, Euphorbia RNAs were reverse transcribed with an oligo–dT primer using an enhanced avian myeloblastosis virus reverse transcriptase enzyme (Sigma-Aldrich).
2.3. PCR Amplification with Hybrid Primers
- Degenerate oligonucleotide primers were designed using the consensus degenerate hybrid oligonucleotide primer (CODEHOP) strategy[39], starting from the alignment of multiple SOD protein sequences. Eight different plant sources were chosen from the GenBank SwissProt database (Ananas comosus accession No.Q9SQL5, Melastoma malabathricum accession No.D5MQD6, Populus trichocarpa accession No.A9PAB4, Gossypium hirsutum accession No. Q3SAX1, Ricinus communis accession No.B9REW9, Gossypium arboreum accession No.B6EBF3, Litchi chinensis accession No. B0FYI8, Zea mays accession No.P23345), and aligned using ClustalW (http://www.ebi. ac.uk/clustalw), and then cut into blocks using the Block Marker software (http://blocks. fhcrc.org/blocks/). Primers were designed using the default parameters at the CODEHOP server (http://blocks.fhcrc.org/codehop.html). Amplification primers for Euphorbia Cu/Zn-SOD cDNA are shown in Table 1. Briefly, the first fragment of partial Cu/Zn-SOD cDNA was obtained using the sense primer 1DF (5'- GCCTGCACGGCTTCcaygtncaygc-3') in association with the antisense primer 1DR (5'-GCCGGTGGTCT TGGACArytcrtgnccnc-3’) Each primer presents the consensus clamp given in upper case whereas the degenerate core is in lower case: y = [C,T]; r = [A,G], and n = [A,G,C,T]. PCR was performed in a solution containing 1.5mM MgCl2, 100mM Tris-HCl, pH 8.3, 50mM KCl, 200mM dNTP mix, 1mM sense primer, 1mM antisense primer, 1μg of Euphorbia cDNA, and 1-3 units of Jump Start AccuTaq LA DNA polymerase mix (Sigma-Aldrich). Thermal cycles of amplification were carried out in a Personal Eppendorf Mastercycler (Eppendorf, Hamburg, Germany) using slightly different programs. The CODEHOP PCR product (400 bp) was sequenced and then used to design specific primers employed in 5'/3' RACE experiments in order to complete the entire coding sequence of the SOD gene.
2.4. Rapid Amplification of cDNA Ends (RACE)
- Rapid amplification of the 5′ end was done using three antisense specific primers (Table 1) and two anchor primers provided in the RACE kit (Roche Diagnostics, Germany) [40]. To perform 5′ RACE, the first antisense specific primer 1R (5’-TTCGTCATCAGGAGCACCATGCTC-3’) was used in a reverse transcription reaction with 2 μg of E. characias total RNA. The first strand cDNA was purified from unincorporated nucleotides and primers by the High Pure PCR purification kit (Roche Diagnostics). A homopolymeric tail was added to the 3’ end of RT-PCR products, and the obtained cDNA was amplified by PCR using the nested antisense-specific primer 2R (5’-TGAGGCCCAGTTGA CATGCAACC-3’) and the oligo(dT)-anchor primer provided in the RACE kit, according to the protocol supplied (Table 1). For 3’ RACE, the first strand cDNA was obtained using the oligo(dT)-anchor primer and amplified using the sense-specific primer 1F (5’-CTGTTGGTGATGATGGCA CTGC-3’) and the antisense PCR anchor primer provided in the RACE kit (Table 1). PCR reactions were performed using 1-3 units of Jump Start AccuTaq LA DNA polymerase mix (Sigma-Aldrich) under different experimental conditions.
|
2.5. Nucleotide Sequence Analysis
- cDNA sequencing was performed with an ABI Prism automated sequencer (Applied Biosystems, Foster City, CA). Nucleotide and deduced amino acid sequence analyses were performed by means of freeware programs. Translation of nucleotide sequences was performed using the ExPASy translate routine software (http://ca.expasy.org/). Similarities were analyzed with the advanced BLAST algorithm, available at the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov/), and with the FASTA algorithm version 3.0 from the European Bioinformatics Institute website (http://www.ebi.ac.uk/fasta33/in- dex). Sequences were aligned with ClustalW. To calculate the isoelectric point (pI) and the molecular weight of the deduced protein we used the ExPasy (http://www.expasy. org/tools/) program.
2.6. Southern Blot Genomic DNA Analysis
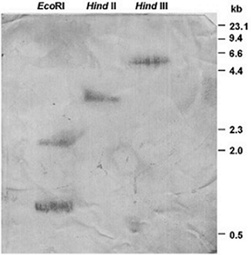
- DNA was obtained from Elx using a Charge Switch Plant DNA Isolation Kit (Invitrogen). Euphorbia genomic DNA (10 µg) was digested with 10-20 units of each EcoRI, HindIII, and HindII (Sigma-Aldrich) in the buffer recommended by the manufacturer, at 37 ℃ for 4 h. Digested DNAs were electrophoresed in 1% agarose gel and transferred overnight to a positively charged nylon membrane (Roche Diagnostics, Mannheim, Germany), by capillary blotting in 20 SSC buffer (3 M NaCl, 0.3 M trisodium citrate, pH 7.0). The blot was baked for 30 minutes at 120 ℃. A probe consisting of a 416 bp PCR amplicon corresponding to nt 80-496 of the Elx SOD cDNA was labeled with digoxigenin-dUTP as described in the DIG System Users Guide (Roche Diagnostics, Mannheim, Germany). Hybridization was performed overnight at 42ºC in Ultrahyb hybridization solution (Ambion) in a probe concentration of 5ng/mL. The membrane was washed twice in 2x SSC, 0.1% SDS for 5 min and twice in 0.1x SSC, 0.1% SDS for 15 min at 42ºC. The DIG-labeled hybridized DNA was detected immunologically by alkaline phosphatase conjugated antibodies in the presence of NBT/BCIP as substrate (Roche Diagnostics). A 0.13-23.1 kb DNA was used as the size marker (Roche Diagnostics).
2.7. Northern Blot Analysis
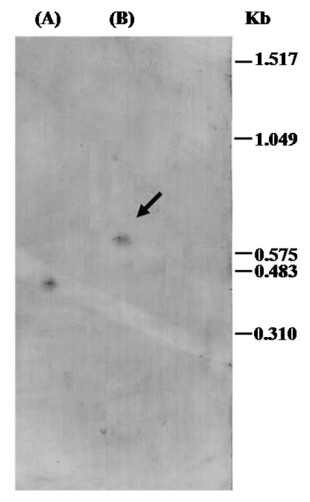
- Ten µg total RNA samples from latex, were dried in a speedvac, dissolved in 20μL of RNA Sample Loading Buffer (Sigma-Aldrich) and heated to 60 ℃ for 15 minutes. The samples were snap-cooled on ice for 5 minutes and then loaded onto a 1% agarose gel containing 0.66 M formaldehyde gel together with an Elx SOD PCR fragment as positive control and 0.31-1.517 kb RNA of the size marker (Roche Diagnostics, Mannheim, Germany). 2µL of 0.2mg/mL ethidium bromide solution was added to the samples before loading onto the gel. The gel was run at 5V/cm in 1×MOPS (20mM MOPS, 1mM EDTA, 5mM sodium acetate, pH 7.0) running buffer and RNA was then transferred overnight to a positively charged nylon membrane (Roche Diagnostics, Mannheim, Germany), by capillary blotting in 20 SSC buffer. Hybridization condition, probe labeling and filter washing steps were identical to those described in the Southern blot analysis section.
2.8. Preparation of Standard Curve for SOD mRNA
- Gene expression of Euphorbia Cu/Zn-SOD in latex during the plant biological cycles was determined by quantitative real-time RT-PCR. The absolute real-time standard curve of the SOD gene was prepared according to the method described by Bustin[41]. ElxSOD cDNA was cloned into the pGEM-T Easy vector (Promega, Madison, WI, USA), and in the presence of SP6 RNA polymerase (Promega), Cu/Zn-SOD RNA was in vitro-transcribed. After transcription, the sample was digested with RNAse-free DNAse and the RNA was quantified with a spectrophotometer. The RNA was reverse transcribed as described above and stored at −20 ˚C until use with quantitative real-time RT-PCR as SOD standard.
2.9. Quantification of SOD Gene Expression by qRT-PCR
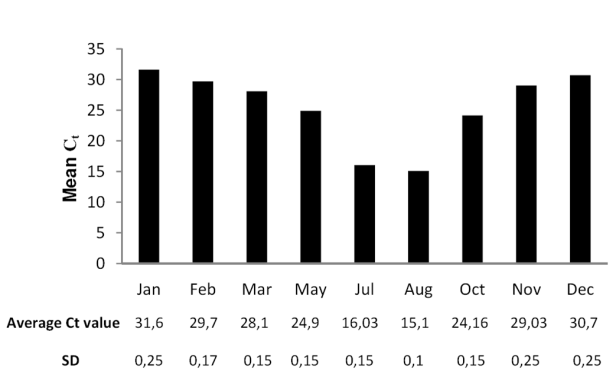
- Real-time quantitative RT-PCR was performed using SYBR Green technology on StepOne™ Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). Primer Express v2.0 software from ABI was employed as the primer design for this purpose. The size of amplicon in the qPCR reaction was 150bp to ensure higher amplification efficiency. The amplifications were performed in a 96-well plate in 25 µL reaction volume containing 12.5µL of 2×SYBR Green Master Mix (Applied Biosystems), 2.5µL of specific Euphorbia SOD cDNA sense SP1 (5’-gcctttctggcctaaaacca-3’) and antisense SP2 (5’- GCAGTACGACCACTAGATCC-3’) primers (10mM), 1µL of template (1µg of Elx cDNA or different concentrations of cDNA SOD standard) and 9 µl of DEPC-water. The thermal profile for SYBR Green real-time RT-PCR was 50 ˚C for 2 min and 95 ˚C for 10 min, followed by 40 cycles at 95 ˚C for 15 s and 60 ˚C for 1 min. In a 96-well plate, each sample was conducted in triplicate. DEPC-water for template replacement was used as negative control. Once the run was finished, the linear relationships between Ct, threshold PCR cycle and the different log concentrations of Cu/Zn-SOD standard, were generated by StepOne™ Real-Time PCR software V2.0 (Applied Biosystems, Foster City, CA) automatically. The different Cu/Zn-SOD Ct values were calculated to actual concentrations of Cu/Zn-SOD mRNA in total RNA based on the SOD standard curve (R2= 0,999). The experiments described above were performed four times at approximately monthly intervals (January, February, March, May, July, August, November, December).
2.10. Statistical Analyses
- Parameters were statistically tested by analyses of variance and comparisons of means were performed with a Duncan test (P < 0.05). Statistical analyses were performed with Statgraphics Plus for Windows, version 5.1 (Statistical Graphics, Englewood Cliffs, NJ, USA).
2.11. Elx SOD PAGE Assay
- The SOD activity was detected using two simple PAGE assays of SOD activity, for positive[42] or negative[43] gel staining. All reagent for SOD PAGE assays were purchased from Sigma-Aldrich. The enzymatic activity was investigated in latex and in the rubber-free acetone powder obtained following the method described by Rinaldi et al.[44], or by the method reported by Freitas et al.[45].
2.12. Nucleotide Sequence Accession Numbers
- The Euphorbia characias Cu/Zn-SOD gene sequences obtained in this study have been deposited in GenBank under Accession Number EF176601
3. Results and Discussion
3.1. Cloning and Analysis of a Cu/Zn-SOD cDNA
- Using the CODEHOP strategy and a progressive primer design strategy, we cloned the cDNA encoding for Euphorbia latex Cu/Zn-SOD. The complete cDNA contains an open reading frame of 462bp corresponding to 152 amino acids encoding a protein with a predicted molecular mass of approximately 15300 Da and a calculated pI of 5,46. The 3′ untranslated region includes two potential polyadenylation signals, respectively 25 and 98 nucleotides downstream from the stop codon, both matching exactly the AATAAA consensus. It seems likely that the second signal represents a near upstream element associated with polyadenylation of the mRNA 3′ end (Figure 1). The sequence for the secretion signal peptide was not detected, which leads us to assume that the SOD gene probably corresponds to a protein located in the cytosol of laticifer cell tissue. The deduced amino acid sequence of Euphorbia SOD compared with those of other plant SODs available from GenBank, showed higher identities (82–84%) with the sequences of cytosolic dimeric Cu/Zn-SODs of other plant species.
3.2. Organization of ElxSOD in the Euphorbia Genome
- In order to determine the copy number of Cu/Zn-SOD gene in Euphorbia characias, the genomic DNA extracted from Elx was restricted with EcoRI, HindIII, and HindII endonucleases and the membrane was exposed to PCR specific fragments of Elx SOD cDNA as a probe. When the genomic DNA was digested with HindII and HindIII, having no internal digestion site within Euphorbia SOD gene, one hybridation signal was detected, whereas EcoRI digestion gave two major bands as expected, since one EcoRI restriction site can be deduced in the cDNA clones (Figure1). The membrane washed under low- and high-stringency conditions showed the same hybrid pattern (Figure 3) suggesting that Euphorbia characias contains a single SOD gene. A similar result was also reported for the genomic organization of maize[50] or Nicotiana plumbaginifolia Cu/Zn-SOD[51], whereas most plants contain multiple gene encoding SOD isoforms[51,53,54].
3.3. Northern Blot Analysis and Quantitative RT-PCR
- Total RNAs from Elx were subjected to northern blotting, using DIG-labeled Euphorbia Cu/Zn-SOD cDNA as a hybridation probe, to investigate the expression of SOD mRNA. Experiments showed a single SOD hybridization band of about 0.7 kb (Figure 4). This finding revealed that homogeneous SOD mRNA population occurred in Elx and that alternative splicing events did not occur. RNAs samples, isolated from latex during the vegetative life of plant, always provided a single hybridation signal confirming that the enzyme arose from one gene producing only one homogeneus population of Euphorbia SOD mRNA. Northern blotting experiments also showed that the gene expression of SOD in latex underwent seasonal fluctuations. Intensive hybridation bands were detectable in RNAs samples isolated and analyzed in July and August, whereas weak signals were detectable when samples where isolated and examined from December to February (not shown). To confirm this result, total RNA was isolated from latex at approximately monthly intervals from January to December and reverse transcribed to cDNAs to perform quantitative RT-PCR (qRT-PCR).
3.4. ElxSOD Enzymatic Assay
- In order to correlate the RNA transcript levels with post-trascriptional events, SOD enzymatic activity was investigated in Elx. We detected the presence of SOD activity in latex by xantine/xantine oxidase/ nitroblue tetrazolium assays in native polyacrilamide gel electrophoresis (PAGE), but without obtaining reproducible quantitative results. Furthermore, the method described by Misra and Fridovich [42], based on the ability of SOD to inhibit the autoxidation of epinephrine at alkaline pH, was ineffective to detect significant amounts of SOD after native PAGE. As previously reported, in Elx are present antioxidant proteins, which are able to remove the H2O2 produced in both assay mixtures. So, we attempted to purify SOD from latex using the method described by Rinaldi et. al.[44], but unfortunately we observed the enzyme precipitation with loss of enzymatic activity after treatment of freeze-dried latex with acetone. Other procedures to obtain latex samples devoid of rubber [45] were not useful.
4. Conclusions
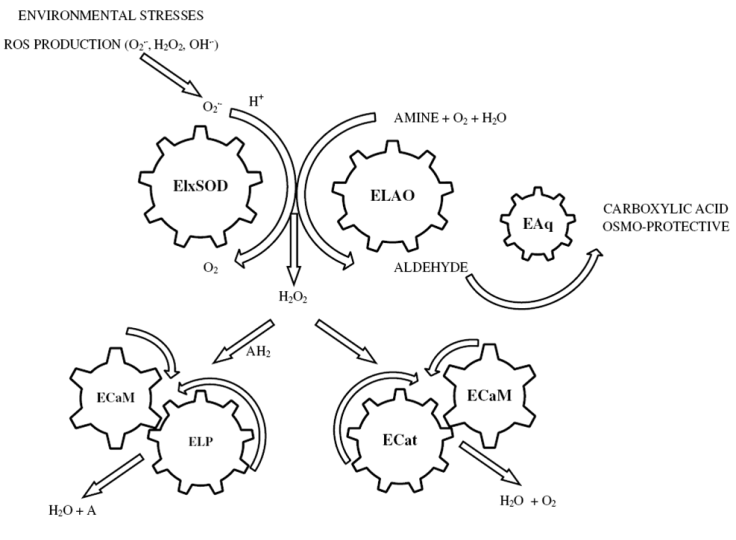
- Both abiotic and biotic stresses may enhance the production of reactive oxygen species (ROS) such as superoxide anion O2–- and H2O2[55,56]. The removal of ROS is essential for the survival of plants during environmental stress. Numerous experimental data demonstrate the possible involvement of laticifer proteins in plant defense mechanisms [57,58]. The molecular studies conducted on Elx revealed the existence of numerous proteins, some of which may be implicated in plant defense mechanisms against invading pathogens/environmental stresses[7,59,11], and in the control of H2O2 homeostasis.Here we demonstrate with molecular biology investigations, that a single SOD isoenzyme occurs in Euphorbia characias and it can help to understand the role that this enzyme plays in the metabolism in vivo. Given the absence of the signal peptide secretion sequence in the SOD cDNA sequence analysis of latex Euphorbia characias, we assume that the putative Cu/Zn-SOD protein is probably located in the cytosol. In this study, we observed that the expression of Cu/Zn SOD is widely induced by dehydration and high temperatures of summer which can be over 40 ° C in Sardinia and in other Mediterranean areas, suggesting that this gene may be involved in the antioxidant defense response under environmental stress. Consequently, molecular characterization of SOD allows us to add a piece to the complicated molecular puzzle previously described[11,60] for E. characias. Theoretically, we could describe the molecular framework of Euphorbia proteins characterized to date, starting from the SOD that catalyze the dismutation of O2–-, produced in response to physiological processes and environmental stresses (Figure 6). The SOD fluctuation expression observed during the seasons, corresponds to a different modulation of transcriptional events, which could be associated with defense responses against environmental stresses. The levels of intracellular H2O2 also result from the reactions catalyzed by the amino oxidase enzyme (ELAO), whose role is to combine the oxidative deamination of biogenic amines, producing the corresponding aldehydes, ammonia (NH3) and hydrogen peroxide[61,62]. At this point, ELP and ECat could locate a metabolic position responsible in the regulation of a large number of H2O2-signaling pathways related to defense against invasion of pathogens or environmental stresses. Both enzymes peroxidase and catalase may be assisted in their action by the intervention of the complex Ca2+/CaM as evidenced by molecular characterization of the two proteins and biochemical experiments conducted on ELP[7]. Finally, the EAq could catalyze the oxidation of aldehydes, potentially toxic metabolites, using them as substrates for the production of osmo-protective cellular molecules, thus indicating swelling of the secretory tissue. Overall, the results from this study together with data obtained in previous studies show that the ability to increase antioxidant system activity in Euphorbia characias latex, might be an important characteristic linked to the drought tolerance in order to limit cellular damage caused by ROS.
ACKNOWLEDGEMENTS
- This study was partially supported by 60% funds from Cagliari University.
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML