-
Paper Information
- Next Paper
- Previous Paper
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
Physical Chemistry
p-ISSN: 2167-7042 e-ISSN: 2167-7069
2013; 3(1): 7-10
doi:10.5923/j.pc.20130301.02
DNA Encapsulation in an Anionic Reverse Micellar Solution of Dioctyl Sodium Sulfosuccinate
Elahe Hosseini Nezhad, Mohammadali Ghorbani, Mehrnoosh Zeinalkhani, Alireza Heidari
Institute for Advanced Studies, Tehran, 14456-63543, Iran
Correspondence to: Alireza Heidari, Institute for Advanced Studies, Tehran, 14456-63543, Iran.
| Email: |  |
Copyright © 2012 Scientific & Academic Publishing. All Rights Reserved.
As nanometric systems, reverse micelles have attracted a great deal of interest especially from the biomedical sciences and widely used in the processes such as the separation and synthesis of biomembranes. We carried out the DNA encapsulation in an anionic reverse micellar solution (RMS) of dioctyl sodium sulfosuccinate (DSS), and studied the interactions through the circular dichroism (CD) and ultraviolet (UV) spectroscopy techniques. The results indicate that the DSS reverse micellar nanosystems provide a suitable medium for encapsulating DNA.
Keywords: CD Spectroscopy, DNA Encapsulation, DSS, RMS, UV Spectroscopy
Cite this paper: Elahe Hosseini Nezhad, Mohammadali Ghorbani, Mehrnoosh Zeinalkhani, Alireza Heidari, DNA Encapsulation in an Anionic Reverse Micellar Solution of Dioctyl Sodium Sulfosuccinate, Physical Chemistry, Vol. 3 No. 1, 2013, pp. 7-10. doi: 10.5923/j.pc.20130301.02.
1. Introduction
- In structural terms, a reverse micellar solution contains clots immediately formed due to the self-assembling of the surfactant molecules in the organic solvents. After 1910, McBain et al. reported the formation of colloidal clots by fatty acid soaps[1-4]. Concerning the quantitative properties and molar conductivity of the soap solutions, McBain came to the conclusion that a part of the soap forms large clots in the aqueous solution called micelle. He found that the micelle formation should be due to the reversible association of positively charged ions. However, this model’s details were revised later. Over the decade 1910s, this idea was considered rather a bold assumption since most researchers at that time believed that the colloidal systems are never thermodynamically stable and their properties depend on the synthesis technique. In 1913, Reychler showed that the sodium cetyl sulfate behaves like the fatty acid soaps and guessed that the molecules are arranged in the micelle such that the hydrophilic core is surrounded by the surfactant hydrophobic end groups[5]. Nowadays, it has been accepted that the clots formed in the ternary mixtures of organic solvent, surfactant, and a very little amount of water are usually spherical with the radius 0.5-5 nm, called the reverse micelle. As evident in Figure 1, such nanostructures are able to solubilize the hydrophilic molecules in the organic phase mass, owing to their aqueous core.
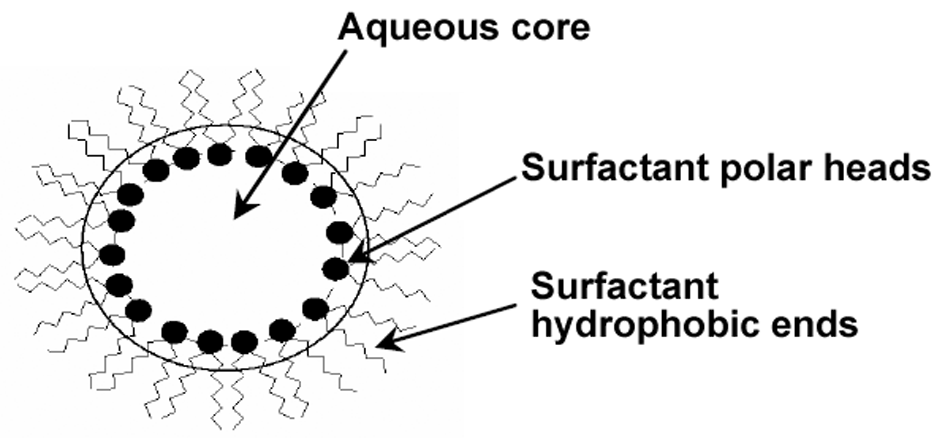 | Figure 1. A schematic view of a reverse micelle structure |
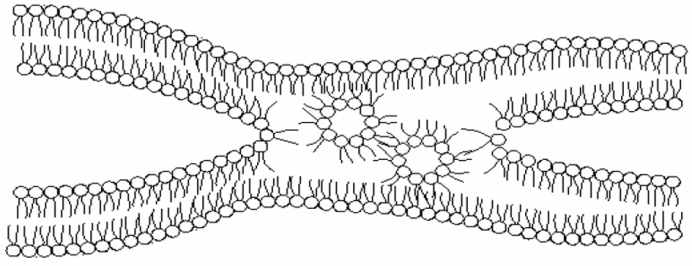 | Figure 2. A schematic view of reverse micelles sandwiched into biomembranes and the intermembrane exchanges |
2. Experimental
- A stochastic ratio of DSS, Sigma Tris-Hydrochloride, lionfish’s DNA, isooctane, ethylenediaminetetraacetic acid (EDTA) was used. The aqueous phase contains 4 mg/mL of DNA and the reverse micellar phase contains 100 mM of DSS. The impurities in the initial RMSs (solid and dust-like suspended particles) were filtered using sterilization membranes (Millex 0.22 m, Millipore, Bedford, MA, USA). The aqueous phase was directly injected at 25℃ to solubilize DNA in the RMS. The concentration of DNA in the RMSs was measured through a Varian Cary 1E UV spectrophotometer at the wavelength 260 nm using a quartz cell with 1 cm path length. The system did not contain DNA in the reference state. Also, the molar ratio of water to surfactant was chosen 18.5 in the reverse micellar phase.The interaction between the reverse micelles and DNA molecules should be investigated after the determination of DNA presence in the reverse micellar phase. In this regard, a circular dichroism (CD) device was employed. This device is an ideal technology to study the structure of the optically active molecules. This optical method is only sensitive to asymmetric molecules and its application in biosystems is of great interest. The CD measurement principles are: adsorption difference (A), levorotatory and dextrorotatory polarized lights that given by the molecules as
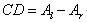 | (1) |
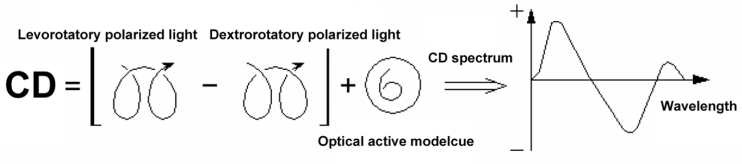 | Figure 3. Circular dichasium (CD) spectroscopy process |
3. Results and Discussion
- Since DNA is a hydrophilic macromolecule, it can only be placed in the aqueous core of the reverse micellar systems. In other words, DNA encapsulation makes its solubilization possible in the organic phase. The UV spectroscopy was utilized to prove DNA solubilization in the reverse micellar phase. Figure 4 shows the UV adsorption of DNA macromolecules in the aqueous and reverse micellar phases. The appearance of the culminating point around the wavelength 260 nm indicates the presence of DNA in these phases. If the conditions are changed and DNA is not encapsulated and directly added to the organic solvent, its natural shape will be immediately changed and the UV spectroscopy shows a relatively horizontal line instead of curve, implying the absorption of the scattered lights.
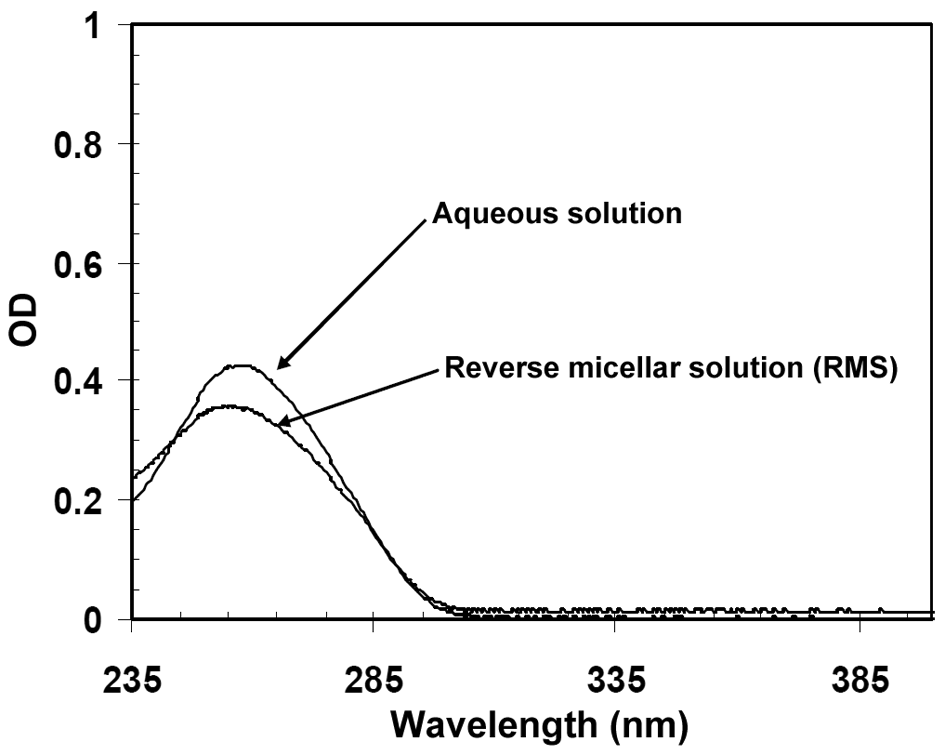 | Figure 4. UV spectrum of DNA macromolecules in the aqueous and reverse micellar phases |
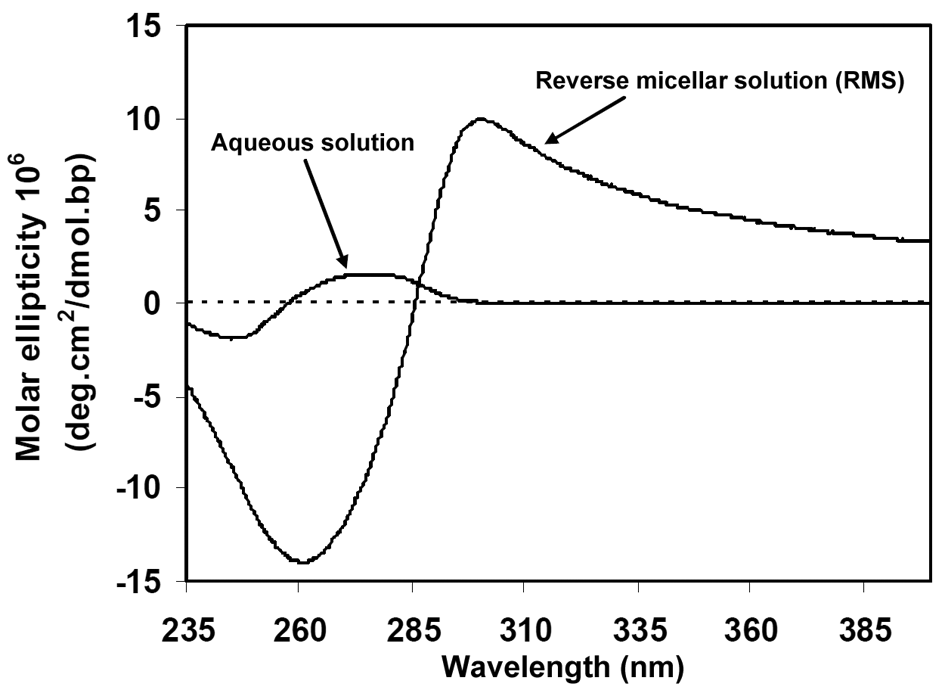 | Figure 5. CD spectrum of DNA macromolecules in the aqueous and reverse micellar phases |
4. Conclusions
- In general, it can be concluded that the reverse micellar system DSS is able to solubilize and condense DNA in the DNA encapsulation process, which is expected to happen naturally in the body.Here in fact a multidisciplinary approach is adopted to produce reverse micellar solution as an in vitro medium to encapsulate DNA for gene delivery. Biophotonics techniques such as CD spectroscopy were employed to optically monitor intracellular trafficking and gene transfection. Generally speaking, the micellar nanochemistry can produce encapsulated DNA molecules, highly mono-dispersed aqueous suspensions of RMS, and surface functionalized by cationic-amino groups.Gel-electrophoresis investigations suggest that the particles can efficiently form a complex with DNA macromolecules, and protect them from enzymatic digestion of DNAse-1 by forming a membrane. DNA and RMS membrane, formed by DSS nanoparticles, are electrostatic bound thanks to the positively charged amino groups, which is detected by appropriately encapsulating DNA and monitoring the surfactant resonance energy transfer between intermembrane and intramembrane DNA macromolecules. CD and UV spectroscopy data clearly show that DNA macromolecules efficiently take up the DSS nanoparticles in vitro, and the DSS nanoparticles surround DNA. CD spectroscopy enabled us to describe the mechanism of DNA encapsulation and gene transfection in RMS media. This investigation suggests that this nanomedicine approach with DSS nanoparticles acting as a drug-delivery platform offers a promising horizon for the targeted therapy techniques and real-time drug reaction monitoring, a part which we aim to further study on in the near future.
ACKNOWLEDGMENTS
- The work described in this paper was fully supported by grants from the Institute for Advanced Studies of Iran. The authors would like to express genuinely and sincerely thanks and appreciated and their gratitude to Institute for Advanced Studies of Iran.
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML