-
Paper Information
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
Research in Otolaryngology
p-ISSN: 2326-1307 e-ISSN: 2326-1323
2014; 3(1): 1-7
doi:10.5923/j.otolaryn.20140301.01
Is Screening for Mitochondrial A1555G Mutation among Assortative Mating Hearing Impaired Families Important? : A Prefatory Quest
Amritkumar Pavithra , Pallikarana Tirumala Harini , Jayasankaran Chandru , Chodisetty Sarvani , Akanksha Rastogi , Mathiyalagan Selvakumari , Mahalingam Subathra , Arabandi Ramesh , C. R. Srikumari Srisailapathy
Department of Genetics, Dr. ALM Post Graduate Institute of Basic Medical Sciences, University of Madras, Taramani Campus, Chennai 600113, India
Correspondence to: C. R. Srikumari Srisailapathy , Department of Genetics, Dr. ALM Post Graduate Institute of Basic Medical Sciences, University of Madras, Taramani Campus, Chennai 600113, India.
| Email: |  |
Copyright © 2012 Scientific & Academic Publishing. All Rights Reserved.
Genetic heterogeneity, multiple phenotypes, consanguinity and marriages between hearing impaired persons have confounded the genetic studies on non-syndromic hearing loss (NSHL). A1555G mutation in the 12SrRNA gene has been identified to be one of the most common mitochondrial mutations and it has been associated with both NSHL as well as aminoglycoside induced ototoxicity. Objective: To screen for the prevalence of mitochondrial A1555G mutation among assortatively mating hearing impaired families from Andhra Pradesh, South India. Materials and methods: Families in which the proband had prelingual, non-syndromic, sensorineural hearing loss and married to a partner who was either of normal hearing status (Deaf x Normal) or was also prelingual hearing impaired (Deaf x Deaf), with at least two generations of family members available for the study were enrolled and genomic DNA was extracted. Mitochondrial A1555G mutation in the 12SrRNA gene was screened by PCR-RFLP method and confirmed by direct sequencing of entire 12SrRNAgene using suitable primers. Additionally, all the individuals carrying the A1555G mutation, along with their family members were screened for GJB2 gene mutations by direct sequencing method. Results: We screened twenty assortatively mating hearing impaired families comprising of one hundred and thirty seven members for A1555G mitochondrial DNA mutation and found seven members in a family with variable phenotypes ranging from normal hearing to moderately severe hearing loss, having this mutation with clear matrilineal transmission. Conclusions: This is the first report from India on the prevalence of A1555G mutation in normal hearing individuals. This study suggests the impending need to screen this common mitochondrial mutation on a large scale not only among the hearing impaired families but also in the normal hearing south Indian population. It would also be worthwhile to include screening for this mutation prior to aminoglycoside treatment in this population to avoid the preventable hearing loss.
Keywords: A1555G mutation, Assortative mating, Hearing loss, India, 12SrRNA
Cite this paper: Amritkumar Pavithra , Pallikarana Tirumala Harini , Jayasankaran Chandru , Chodisetty Sarvani , Akanksha Rastogi , Mathiyalagan Selvakumari , Mahalingam Subathra , Arabandi Ramesh , C. R. Srikumari Srisailapathy , Is Screening for Mitochondrial A1555G Mutation among Assortative Mating Hearing Impaired Families Important? : A Prefatory Quest, Research in Otolaryngology, Vol. 3 No. 1, 2014, pp. 1-7. doi: 10.5923/j.otolaryn.20140301.01.
Article Outline
1. Introduction
- Profound hearing loss is the most frequent sensory disorder affecting at least one in 500 newborns worldwide[1]. Hearing loss (HL) is genetically heterogeneous and has variety of clinical presentations. It is estimated that over 50% of HL has genetic predisposition, while 20-25% is due to identifiable environmental factors which include prenatal or perinatal viral infections, especially cytomegalovirus infection, acoustic or cerebral trauma leading to cochlear damage or ototoxic drugs such as antibiotics, and the remaining 25-30% of HL is of unknown etiology. Among the genetic causes, autosomal recessive inheritance accounts for about 80% of HL, followed by about 20% due to autosomal dominant inheritance, while X-linked inheritance and mitochondrial inheritance contribute to about 1% of HL each[2-4].Several mutations in the mitochondrial DNA (mtDNA) have been found to be associated with syndromic, non-syndromic and aminoglycoside-induced hearing loss. The role of mtDNA mutations in non-syndromic hearing loss (NSHL) is either as a primary or as a predisposing factor. Hearing loss can be due to inherited, acquired, heteroplasmic or homoplasmic mtDNA mutations[5]. According to MITOMAP[6], till date, the pathogenicity of four mt DNA mutations has been clearly established in NSHL: C1494T and A1555G, located in the 12S ribosomal RNA gene (MTRNR1, MIM 561000) and A7445G and T7511C, located in the ser (UCN) transfer RNAgene (MTTS1, MIM 590080). Among the MTRNR1 mutations, C1494T and A1555G are associated with non-syndromic and aminoglycoside induced HL[7]. These MTRNR1 mutations are presumed to alter the secondary structure of the 12SrRNA molecule so that it resembles closely its bacterial counterpart, the 16SrRNA molecule, leading to greater aminoglycoside susceptibility which results in destruction of the sensory hair cells in the inner ear that are associated with auditory function[8]. As aminoglycosides target the bacterial 16S rRNA molecule, the cumulative effect of MTRNR1 mutations and use of aminoglycosides can be correlated[5]. However, the mechanism of action for hearing loss in the absence of exposure to aminoglycosides is unclear. A1555G mutation, in which there is a substitution of Adenine to Guanine at the 1555 position, has been found in more than 120 families across the world, making it the most common genetic factor for HL amongst the identified ones and also provides an example on how environmental factors can interact with genetic predisposition[9, 10]. The A1555G mutation in the 12SrRNA gene was first described in a large Arab-Israeli pedigree[11], and subsequently found in many families of various ethnic backgrounds[12-16]. The incidence of A1555G mutation in various populations with non-syndromic hearing loss and aminoglycoside ototoxicity is comparatively higher than those without aminoglycoside exposure[17]. In the absence of aminoglycoside exposure, A1555G mutation can present a host of phenotypes ranging from profound hearing loss to moderate hearing loss to even apparently normal hearing between families as well as within families[5, 11, 18]. Mutations in the nuclear genes GJB2 and TRMU are reported to influence the degree of hearing loss in patients with the A1555G mutation[19, 20]. Further, many mitochondrial tRNA variants have also been reported to modulate the penetrance of the A1555G mutation[20, 21].Although there has been several studies on the relationship between mitochondrial mutations and non-syndromic hearing loss across the world[12-16, 22], including India [23, 24] there has not been any report to our knowledge, on its prevalence in assortatively mating Hearing Impaired(HI) mates and their families from any part of the world. With a trend of marriages between HI persons becoming increasingly common, there is a pressing need to understand the mutation dynamics in such settings. Our present study was aimed at understanding the contribution of A1555G mt DNA mutation to non-syndromic deafness in a special cohort of HI mates and their families from India using a comprehensive approach of screening the HI mates and their extended families.
2. Method
- This study was approved by the Human Ethical Committee, University of Madras.SubjectsWe identified hearing impaired (HI) mates by approaching various adult organizations and associations for the HI in Andhra Pradesh, south India. We included only those families in which the proband was prelingual HI and married to a partner who was either of normal hearing status (Deaf x Normal, or DXN in short) or was also prelingual HI (Deaf x Deaf, or DXD in short), with at least two generations of family members available, for the study. Written informed consent was obtained from all participants in every family prior to their participation in the study, in accordance with the Institute Review Board and Ethics Committee, University of Madras. Detailed family pedigrees were drawn. Information on consanguinity, age at onset of HL, detailed prenatal and perinatal history, use of ototoxic drugs (aminoglycosides), etc. was obtained through a detailed questionnaire. The HI probands were evaluated by pure tone audiometry (PTA) using Graphic Digital RS-1 Speech Audiometer. Where both the mates were HI, information was obtained from at least two speaking relativeor with the help of a sign language expert in our team. Twenty assortatively mating HI families(Seven DXD families and Thirteen DXN families) with 137 members in all were screened during this study. All the available family members in each of these families, irrespective of their hearing status, were included in the screening procedure along with the proband owing to the variable phenotypic expressivity of this mutation. Eight milliliters of blood was drawn from each member and was processed further for A1555G mutation screening.Molecular AnalysisA part of 12SrRNA gene harboring the A1555G mutation region was amplified by PCR using forward primer: 5’ AGA AAT GGG CTA CAT TTT CTA CCC 3’ corresponding to position 1354-1377 and reverse primer: 5’ GTT CGT CCA AGT GCA CTT TCC A 3’corresponding to position 1580-1601 in the mtDNA[25]. The presence of A1555G mutation was detected by restriction digestion of PCR products (248bp)using BsmA1 enzyme (New England Bio Labs®) and checked using 3% agarose gel electrophoresis. The expected size post-restriction digestion, in the absence of mutation was 197bp and 51bp. Substitution of A to G at 1555 position abolishes the restriction site yielding a single band of 248 bp size.The presence of the A1555G mutation was further confirmed by sequencing the entire 12SrRNA gene using the primer pairs 12SF-5’TTTAGACGGGCTCACATC 3’ and 12SR-5’CTCTGGTTCGTCCAAGTG 3’[7] and ABI Prism Big Dye Terminator Cycle sequencing reaction kit (Applied Biosystems) on an ABI 3730 Automated Sequencer. The resultant sequences were compared with the ‘rCRS: Revised Cambridge Reference Sequence NC_012920’[26], to identify the nucleotide changes. Additionally, all the individuals carrying the A1555G mutation, along with their family members were screened for GJB2 gene mutations by direct sequencing method, previously described by Ramshanker et al.[27].
3. Results
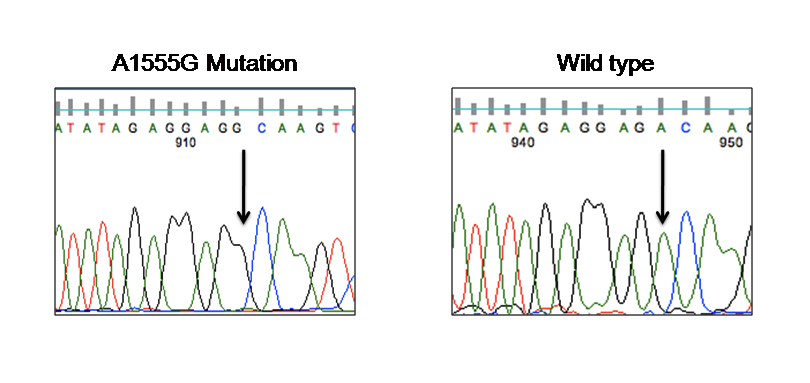
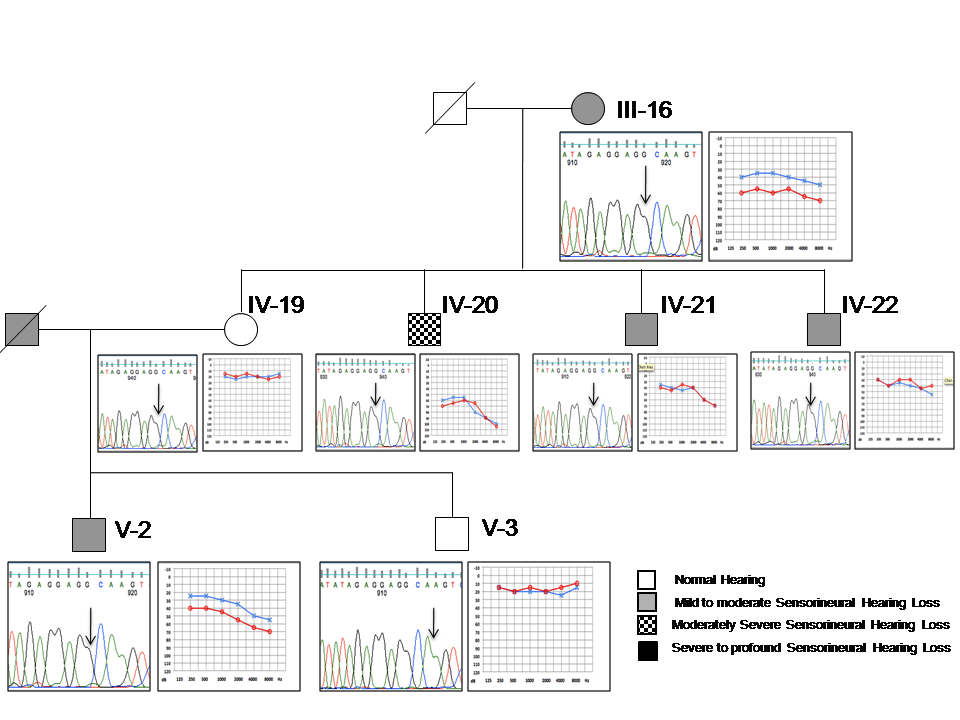
- Among the 20 assortatively mating HI families screened for the A1555G mutation, one family (DXN NLR12) where the HI husband is married to a normal hearing woman showed the presence of this mutation (Fig.1). Seven extended family members (III-16, IV-19, IV-20, IV-21, IV-22, V-2 &V-3) of DXN NLR12 family spread in three generations showed the presence of A1555G mutation by PCR-RFLP analysis. The presence of homoplasmic A1555G mutation was further confirmed by direct sequencing of the whole mitochondrial 12SrRNA gene (Fig. 2 and 3). No other mitochondrial mutation or variant was observed in these seven A1555G positive individuals as well as their family members. Direct sequencing of GJB2 gene showed the absence of mutations, variants and polymorphisms in all the members of this family.
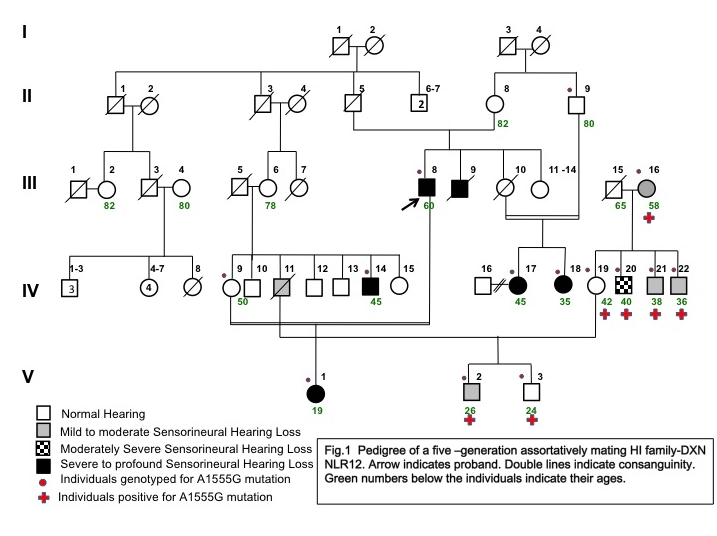 | Figure 1. Pedigree of five-generation assortatively mating Hearing Impaired family- DXN NLR 12 |
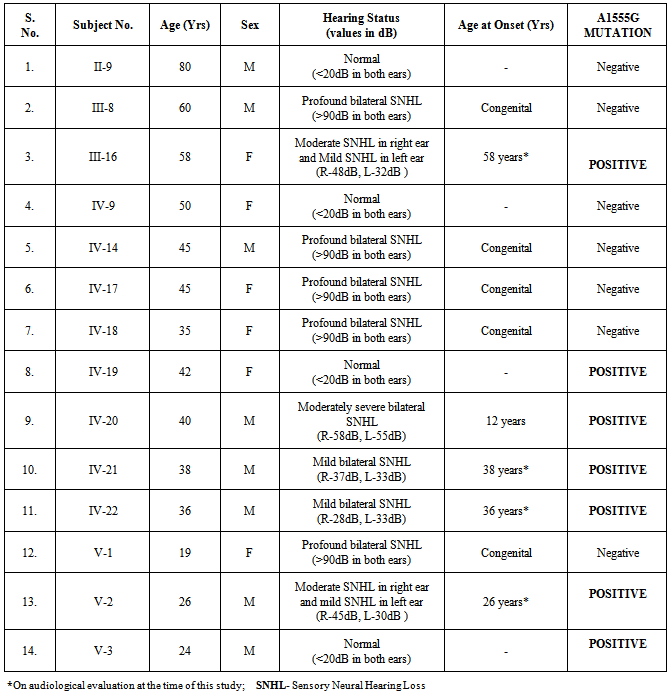
- In this family, five members had prelingual profound bilateral sensorineural hearing loss (SNHL) including the proband (III-8), five members showed variable levels of hearing loss and four members had normal hearing status. Clinical characterization has been presented in Table 1. Four individuals who self-reported to be normal hearing showed mild to moderate hearing losses on our clinical evaluation by PTA. Their audiological profiles are: III-16 and V-2 showed moderate SNHL in the right ear and mild SNHL in the left ear; IV-21 and IV-22 showed mild SNHL (Fig. 3). There were no available recent medical prescriptions or detailed medical records with them to verify the usage of aminoglycoside antibiotics. These individuals were also not able to associate their variable hearing losses to any recent medication. On the other hand, IV-20 who had a history of difficulty in hearing since a bout of fever around 12 years of age, showed bilateral moderately severe SNHL on our evaluation.
|
4. Discussion
- Marriages between the hearing impaired (HI) are well recognized and may represent one of the most common genetic traits in which an altered mating structure occurs in the human populations[4]. In fact, there are very few other genetic traits for which such a high degree of assortative mating has been described. Assortative mating within a population can have profound influence on the distribution of genes in that population[28, 29].In the present study, twenty assortative mating HI families (Seven DXD families and Thirteen DXN families) from Andhra Pradesh, south India, were screened for the presence of mitochondrial A1555G mutation. Extended family members (seven members spread over three generations with variable status of hearing) from a five-generation DXN mating HI family showed the presence of A1555G mutation. The inheritance pattern showed clear matrilineal transmission with A1555G positive mother passing toher four offsprings, out of which the female offspring further transmitted the mutation to the subsequent generation. All these seven members have shown variable degrees of hearing losses on our audiometric evaluation, ranging from normal hearing to moderately severe hearing loss. The only person (IV-20) who had early onset moderately severe hearing loss post a fever bout and medicationdid not have any medical documentation. Varying degrees of penetrance and severity of HI has been previously described in many studies pointing to the fact that A1555G mutation by itself is not sufficient enough to produce the deafness phenotype [7, 11, 30]. It has been suggested that certain nuclear modifiers such as 35delG mutation in the GJB2 gene and A10S mutation in TRMU gene, certain mitochondrial haplogroups and presence of aminoglycosides modulate the phenotypic expression of the A1555G mutation[31, 32]. While the role of other mutations in the mitochondrial 12SrRNA gene and GJB2 gene mutations has been excluded in these positive individuals, variable phenotypic expression could be attributed to one of the many other factors previously described. It should also be noted that the severity of hearing loss is only up to moderately severe range, that too in one individual, while others have losses in the mild to moderate range which could be attributed to environmental causes such as noise.Identification of two normal hearing individuals harboring the A1555G mutation has important medical implications as these family members are at increased risk of developing aminoglycoside induced hearing loss. Aminoglycosides are a class of antibiotics that are highly effective in the treatment of life threatening Gram-negative bacterial infections such as meningitis and bacterial sepsis in infants. They are widely used in developing nations like China, India and many other Asian countries as well as South African countries as first line of drugs, especially Streptomycin, to treat Multi-Drug Resistant Tuberculosis owing to their broad spectrum antimicrobial activity and cost-effectiveness[12, 33]. These drugs act by binding directly with the bacterial 16S ribosomal RNA subunit and result in mistranslation or premature termination of the bacterial protein synthesis. However, aminoglycosides also induce tissue-specific toxicity in cochlea and kidneys. These drugs are administered in doses based on their body weight and their toxicity is dose-related. Individuals receiving sufficiently high doses suffer from functional and/or morphological damage in the cochlea. As aminoglycosides are cleared more slowly from inner ear fluids than from serum, the toxic effects are more pronounced leading to the irreversible death of outer hair cells in the organ of corti, predominantly at the basal turn of the cochlea[8]. A1555G mutation in the MTRNR1 gene which codes for the mitochondrial 12SrRNA brings about conformational changes at the A site of 12SrRNA, which more closely resembles the bacterial 16SrRNA counterpart, leading to increased affinity to the binding of the aminoglycosides leading to ototoxicity, even at lower dosage levels[34, 35]. All the A1555G positive individuals in our study hence were informed of their high-risk status for aminoglycoside induced hearing loss. A detailed genetic testing report that includes the list of antibiotics to be avoided was given to each one of them and was appropriately counseled about the risks associated with aminoglycoside medication. They were also advised to discuss this report with their family physician in the future event of a need for aminoglycoside administration. There have been two studies on the prevalence of A1555G mutation among the Indian HI individuals till date. A study on 303 HI individuals by Padma et al (2012) reported three sporadic individuals with profound hearing loss harboring the A1555G mutation and none in the controls, reporting a frequency of 1.0% for Indian patients. Another study by Bhalla et al (2009) among the North Indian HI individuals had shown absence of this mutation.Although in our present study we did not find A1555G mutation among the HI probands, we have noted this mutation among seven extended family members spread over three generations, due to our not restricting the screening to just the HI probands. The presence of this mutation among the normal hearing individuals stresses the importance of finding out the prevalence of this mutation on larger population, keeping in mind the variable phenotype exhibited by this mutation.
5. Conclusions
- Identification of this mutation has a dramatic impact on the medical care as knowledge of presence of A1555G mutation in apparently normal hearing individuals can potentially avert developing hearing impairment due to aminoglycoside treatment. It would hence be worthwhile to screen for the presence of mutations in the 12SrRNA gene, especially the A1555G mutation in this population with higher frequency of this mutation, prior to administration of aminoglycoside antibiotics to avoid preventable hearing loss.
ACKNOWLEDGEMENTS
- We are grateful to all the families for their participation. This study was supported by UGC-MAJOR GRANT and UICIC Program of University of Madras to CRS.
Abbreviations
- HL, hearing loss; NSHL, non-syndromic hearing loss; mtDNA, mitochondrial DNA; HI, hearing impaired.
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML