-
Paper Information
- Next Paper
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
Nanoscience and Nanotechnology
p-ISSN: 2163-257X e-ISSN: 2163-2588
2015; 5(3): 45-56
doi:10.5923/j.nn.20150503.01
Evaluation of Callus Responses of Solanum nigrum L. Exposed to Biologically Synthesized Silver Nanoparticles
Emad A. Ewais1, Said A. Desouky2, Ezzat H. Elshazly2
1Botany and Microbiology Department, Faculty of Science, Al-Azhar University, Cairo, Egypt
2Department of Botany, Faculty of Science, Al-Azhar University, Assiut, Egypt
Correspondence to: Emad A. Ewais, Botany and Microbiology Department, Faculty of Science, Al-Azhar University, Cairo, Egypt.
| Email: |  |
Copyright © 2015 Scientific & Academic Publishing. All Rights Reserved.
Effect of biosynthesized silver nanoparticles (AgNPs) on growth, anatomy, and protein and DNA of Solanum nigrum callus was investigated in vitro. Three concentrations of aqueous silver nanoparticles conjugated with camel milk were used in the present study. Murashige and Skoog (MS) nutrient media supplemented with different concentrations of AgNPs, 0, 2, 4 and 8 mg/l, were evaluated for their effects on the callus induction from the leaf explants of Solanum nigrum. Compact calli with white and greenish colour were obtained after 10 days upon control culture (no AgNPs), whereas friable watery calli with white, greenish or yellowish colour were observed after 10-13 days upon culture supplemented with AgNPs. Interestingly, it was found that exposure to AgNPs increased callus fresh weight and the intensity of callus formation. These were associated with the deformity of callus morphology. Light microscopy observations showed that the nanoparticles damaged cell wall. This was clear with the high concentration of AgNPs. Variation in protein pattern were recorded between callus control (no AgNPs) and callus exposure to AgNPs. Genetic stability between callus control and callus under AgNPs exposure was interrelated.
Keywords: Solanum nigrum, Silver nanoparticles, Callus anatomy, Protein and genetic stability
Cite this paper: Emad A. Ewais, Said A. Desouky, Ezzat H. Elshazly, Evaluation of Callus Responses of Solanum nigrum L. Exposed to Biologically Synthesized Silver Nanoparticles, Nanoscience and Nanotechnology, Vol. 5 No. 3, 2015, pp. 45-56. doi: 10.5923/j.nn.20150503.01.
Article Outline
1. Introduction
- The field of nanotechnology is one of the most active areas of research in modern material science. Nanoparticles are being considered to be the fundamental building blocks of nanotechnology. Nanotechnology is interdisciplinary which includes physics, chemistry, biology, material science and medicine. Instead of using toxic chemicals for the reduction and stabilisation of metallic nanoparticles, the use of various biological entities has received considerable attention in the field of nanobiotechnology [1]. Biological methods are regarded as safe, cost-effective, sustainable and environment friendly processes for the synthesis of nanoparticles [2]. Silver nanoparticles have been successfully synthesized using various bacteria [3], fungi [4] and plants [5].Nanoparticles are becoming an area of research interest due to their unique properties, such as having increased electrical conductivity, ductility, toughness, and formability of ceramics, increasing the hardness and strength of metals and alloys, and by increasing the luminescent efficiency of semiconductors [6]. Plants are an essential base component of all ecosystems and play a critical role in the fate and transport of AgNPs in the environment through plant uptake and bioaccumulation [7]. Silver nanoparticles (AgNPs) are currently one of the most widely commercially used nanomaterials [8]. The risk of nanomaterials usage represents their possible accumulation in the environment with subsequent entry into the food chain, which is closely connected with their accumulation in organisms [9]. Plants represent an important trophic level [10]. However, there are only limited studies focused on the effect of nanomaterials on plants. The first point of possible phytotoxicity consists in interactions between nanomaterials and soil components and microorganisms, which significantly modify uptake of nutrients by plants [11]. The second point is closely connected with the uptake of nanomaterials (root, foliar), and their accumulation and transport within plant body with subsequent interactions with biomolecules, such as nucleic acids, proteins including enzymes, and cell structures, such as cell walls and biomembranes [12]. In particular the cell wall of plant cells represents a crucial structure in nanoparticle uptake compared to animal cells [13]. Both positive and negative effects on plants have been demonstrated. Lu et al. demonstrated both positive and negative effects of nano-SiO2 and nano-TiO2 on nitrate reductase in soybean [14]. Enhancement of biomass production in spinach (Spinacia oleracea) after application of TiO2 nanoparticles has been recorded in a study by Gao et al., [15].An understanding of the interactions between nanoparticles and biological systems is of significant interest. Studies aimed at correlating the plant cells responses such as morphological, anatomy, physiology and genetic stability at molecular level are under way. These fundamental studies will provide a foundation for understanding the interaction of nanoparticles with biological systems. So, in this study, we employed callus culture of Solanum nigrum as a model species to investigate the effect of biosynthesised silver nanoparticles (AgNPs) prepared using camel milk on some morphological and anatomy characters of callus and assessment of genetic stability at protein and DNA levels.
2. Materials and Methods
2.1. Plant Material
- Seeds of Solanum nigrum L. were obtained from Botany & Microbiology Dept., Faculty of Science, Al-Azhar Univeristy, Assuit, Egypt.
2.2. Seeds Sterilization Using NaOCl 5.25%
- Seeds In vitro germination using basal MS medium [16] containing 3% sucrose and callus induction from developing leaf explant using 3.0mg/l NAA and 5.0mg/l BA were done as described previously [17].
2.3. Synthesis of Silver Nanoparticles
- Silver nitrate was purchased from Sigma-Aldrich (St. Louis, MO). Camel milk was procured from local market. Camel milk (4 mL) was mixed with 96 mL of 1 mM silver nitrate solution and the resulting mixture was incubated for 8 h in rotatory shaker (180 rpm) at room temperature according to the method described by Lee et al. [18]. Reduction of silver ions in the reaction mixture was monitored by change in colour of the reaction mixture from milky white to dark brown. The obtained nanomaterial diffusion) was freeze dried at -80°C and used for characterization studies. Autoclaved water was used as a control negative for this experiment. The surface morphologies and sizes of the AgNPs were examined using biological transmission electron microscopy (JEOL JEM 2010 transmission electron microscope) and X-ray diffractometer as Moharram et al. [19].
2.4. Longitudinal Section of Callus and Observation under Light Microscope
- For anatomical study small portions of calls of all samples with age thirty days was fixed in FAA (formalin: acetic acid: 70% ethanol; 1: 1: 18, v/v) for 48 hrs. at room temperature. Following a standard dehydration in a graded ethanol series, samples were washed in changes of ethanol 70% to 99.9% ethanol with a difference of 10% in concentration and three hrs. difference in time ate the first two changes and then two hrs. difference between the rest changes. Then sample clearing was carried by means of ascending concentrations of ethanol-xylene with concentrations 10% to 100%. Then samples were embedded in paraffin wax at 45-55ºC for 48 hrs. with changing paraffin wax each 12 hrs. [20]. Longitudinal sections, 12-15 µm thick, were cut with steel blade on IHC World KD-1508A rotary microtome, then sections were fixed on glass slides by Haupt’s adhesive (1gm gelatine powder, 100 ml warm distilled water[note: soluble gelatine should be filtrated before glycerol add, 7.5 ml glycerol and small phenol crystal) [21]. Before sections stalk on slides they were gently heated on hotplate at 30-35°C. After minimum 12hr. the fixed sections were stained with Safranin O-Fast-green double stain. Safranin O solution prepared by dissolving 1gm of stain powder in100 ml ethanol 50% while Fast-green solution prepared by dissolving 1gm of stain powder in100 ml ethanol 95%, then stains solutions filtrated. After staining, sections mounted in Canada balsam [22]. Staining chart (NO.) shows: stage A) de-waxing, stage B) rehydration for Safranin O stain, stage C) dehydration for Fast-green stain and stage D) for loading sections with Canada balsam.Observation and photomicrographs were achieved using XSZ-N107 Research Microscope fitted with Premiere MA88-900 digital camera Staining Chart (NO.) (Safranin O - Fast green).
2.5. Protein Electrophoresis (SDS-PAGE)
- Total protein was extracted from callus control (no AgNPs) and callus exposure to different concentrations of AgNPs. Then, it was ground to fine powder with pestle and mortar. Ten mg of powered flour was homogenized thoroughly with 400 μl extraction buffer using vortex. The extraction buffer was prepared by dissolving 0.6 g Tris base, 0.2 g Sodium Dodecyl Sulfate (SDS) and 30 g of urea in 50 ml of double distilled water. One ml of β- mercaptoethanol was added and then the solution was diluted to 100 ml with double distilled water. The mix was kept overnight at 4°C and then centrifuged at 13000 rpm for 10 minutes at room temperature (Sigma 3K 18 Bench Top centrifuge). For qualitative analysis of protein, 20 μl of the extracted protein was boiled in a water bath for 3-5 min and loaded on Sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE) containing 12.5% resolving gel and 4% stacking gel [23] using 3 μl bromophenol blue as tracking dye. The samples were then loaded in equal amounts (15 μl) and a molecular weight marker standards (5 μl) (Prism Ultra Protein Ladder) were loaded at on to each well. Electrophoresis was carried out at 150 V and 25 mA until tracking dye reached to the bottom of the gel. The gels were resolving gel and 4% stacking gel (Lammeli, 1970) using 3 μl bromophenol blue as tracking dye. The samples were then loaded in equal amounts (15 μl) and a molecular weight marker standards (5 μl) (Prism Ultra Protein Ladder) were loaded at on to each well. Electrophoresis was carried out at 150 V and 25 mA until tracking dye reached to the bottom of the gel. The gels were destained in solvent composed of 40 ml of methanol, 10 ml glacial acetic acid and 50 ml distilled water. The gel was stained overnight in 25 ml of Coomassie Brilliant Blue (R-250) staining buffer. The gels were photographed and the molecular weights of the polypeptide bands were estimated by correlating position of the molecular weight marker standards.
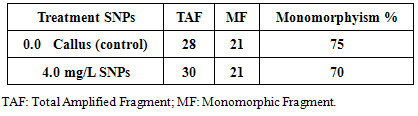
2.6. Random Amplified Polymorphic DNA (RAPD) Analysis
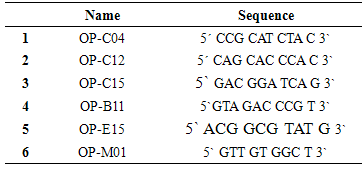
- Approximately 100 mg of fresh callus (callus control, no AgNPs, or callus exposure to 4mg/l AgNPs) were immersed in liquid nitrogen for DNA extraction using DNeasy plant Mini Kit (QIAGEN Hilden, Germany). PCR reaction was performed in 30μl volume tubes contained dNTPs (2.5 mM), MgCl2 (25 mM), 3 μl Buffer, 2 μl Primer, 0.20 μlTaq DNA polymerase (5U/μl), 2 μl Template DNA and 16.80 μl distilled H2O in an automated thermal cycle (Techno 512) programmed for one cycle at 94°C for 4 min followed by 45 cycles of 1 min at 94°C, 1 min at 57°C, and 2 min at 72°C. The reaction was finally stored at 72°C for 10 min. Based on the clear and accurate amplified bands profiles, different six primers were selected (Table 1). The reaction products were analyzed by electrophoresis on 1.2% agarose gels, stained with ethidium bromide and photographed under UV transilluminator by digital camera with UV filter adaptor. The DNA ladder 100 bp (Pharmacia) was employed as molecular markers for bands molecular size. Each amplified band profile was defined by the presence or absence of bands at particular positions on the gel.
|
3. Results and Discussion
3.1. Biosynthesized AgNPs
- The present study is an attempt to synthesize AgNPs using camel milk. The colour of reaction mixture turned from milky white to dark brown after exposed to sunlight for 30 min of reaction, indicating AgNO3 reduction. The intensity of the brown colour increased after 15 mint of incubation, giving it a darker look (Fig. 1). Scanning electron microscopy dispersive spectroscopy peak, approximately at 3ke V, confirmed the presence of silver nanoparticles. Scanning electron microscopy showed that nanoparticles are mostly circular with an average size of 30-90 nm (Fig. 2).
 | Figure (1). Showing colour changes (a) Camel milk before treatment; (b) camel milk after treatment with AgNO3 showing brownish colour |
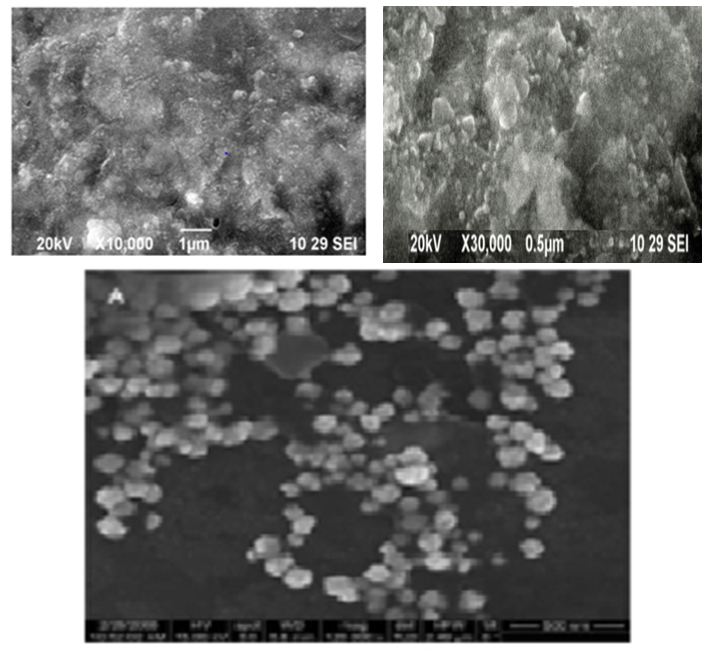 | Figure (2). Particles are mostly circular in shape with the average size of 30–90 nm by using scanning electron microscope |
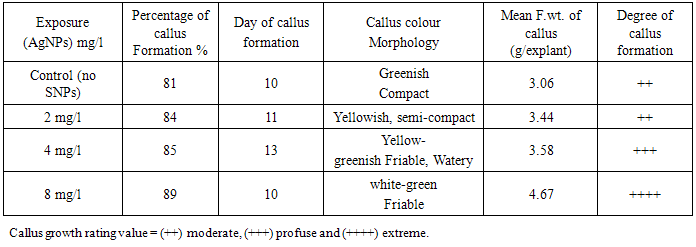
3.2. Effect of Silver Nanoparticles (AgNPs) on Callus Growth
- The results showed that AgNPs had a stimulating effect callus formation, while low and medium concentrations delayed the day of callus formation. It was observed that with increase AgNPs concentration, the degree of callus formation also increase (Table 2). In this respect, it was reported that the AgNPs concentration of 20, 40 and 60 ppm showed statically significant stimulation on shoot and root elongation of common bean and corn [26].
|
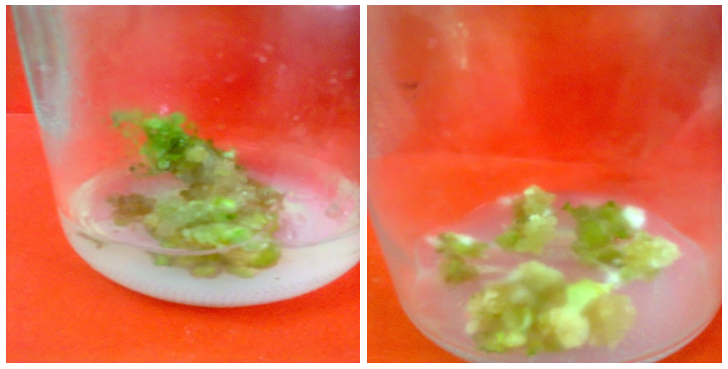 | Figure (3). Compact callus with nodular structures from Solanum nigrum explants cultured MS control medium (with no AgNPs) |
 | Figure (4). Semi-compact callus from Solanum nigrum explants cultured on MS medium supplemented with 2 mg/l AgNPs |
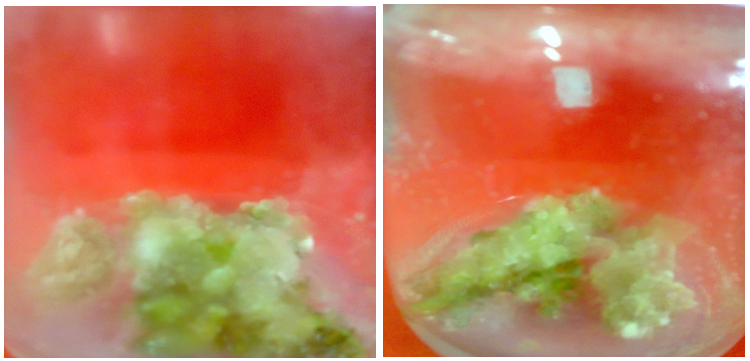 | Figure (5). Friable callus from Solanum nigrum explants cultured on MS medium supplemented with 4 mg/l AgNPs |
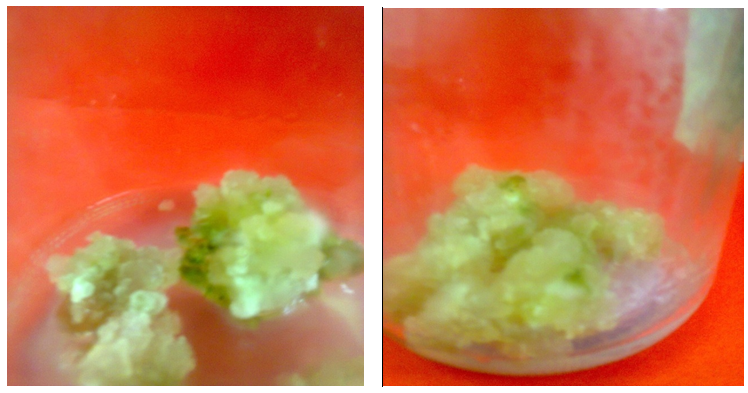 | Figure (6). Friable, greenish white, callus from Solanum nigrum explants cultured on MS medium supplemented with 8 mg/l AgNPs |
3.3. Effect of Silver Nanoparticles (AgNPs) on Callus Anatomy
- It was observed (Fig. 7) that the deformity of callus structure of Solanum nigrum exposure to media containing AgNPs nanoparticles as compared to media without AgNPs (control). It was noticed that no damage for cell wall in callus of control (A), while damages were found in cell wall of callus exposure to AgNPs 2mg/l (B) and with increase in AgNPs, 4mg/l (C), the damage of cell wall also increase and it was dramatically with 8mg/l (D). These results coincide with those Harajyoti and Ahmed [28] who studied the phytotoxicity of silver nanoparticles solutions on Oryza sativa. They found that these nanoparticles had caused adverse effect on damage to external and internal portions of cells and breakage in cell wall of the roots. They added that the entry of particles is clearly seen by breaking the vacuoles. Accumulations of nanoparticles and its toxic effects on test plant species is totally depend upon the concentrations and exposure time.
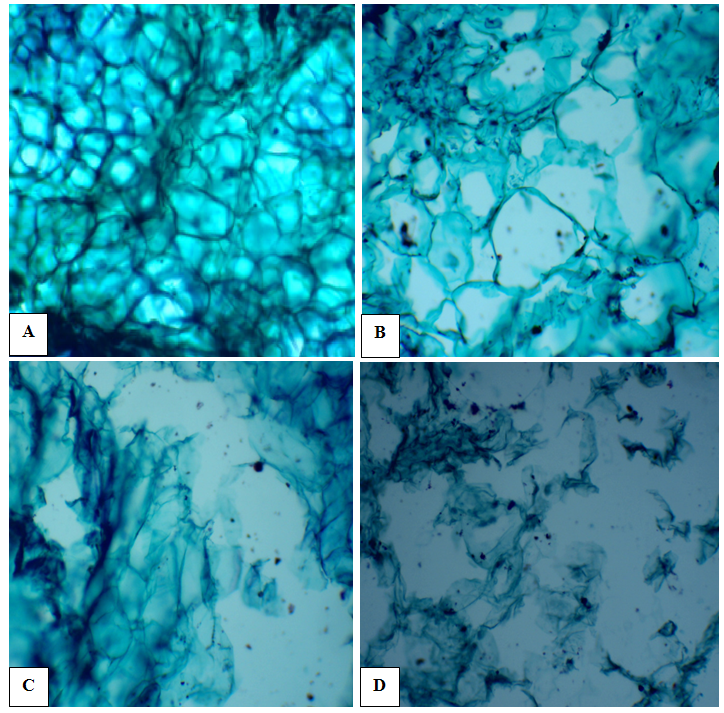 | Figure (7). Light microscopic observation of longitudinal sections of callus of Solanum nigrum L. under treatments of control (A); 2 mg/L AgNPs (B); 4mg/L AgNPs (C) and 8mg/L AgNPs(D). (X100) |
3.4. Assessment of Genetic Stability at Protein Level through SDS-PAGE Biochemical Marker
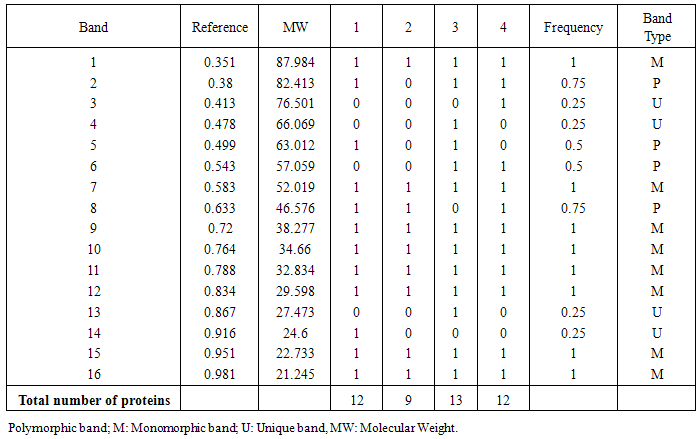
- The protein profile system of Solanum nigrum revealed the biochemical variation and evolutionary relationship among callus control (no AgNPs) and callus under the treatment of AgNPs (2, 4 and 8mg/l) was demonstrated in figure (8). The molecular weights of detected bands for all samples ranged from 21 to 88 KDa. There were 12 bands detected at molecular weight 88, 82, 63, 52, 47, 38, 35, 33, 30, 25, 23, 21 KDa in the callus control (no AgNPs). Only 9 bands were detected in lane 2 (callus treated with 2mg/l AgNPs), whereas 3 bands with molecular weight 83, 63 and 25 KDa were absent. Callus treated with 4mg/l showed 13 bands, with two new bands at molecular weights at 66 and 27 KDa, in addition to disappear one band with molecular weight 47 KDa relative to that observed to control callus. Furthermore, Land 4 (callus treated with 8mg/l SNPs) showed the appearance of two new bands (77 and 57 KDa) and absence of two bands (66 and 63 KDa) as compared to control callus (Table 2).
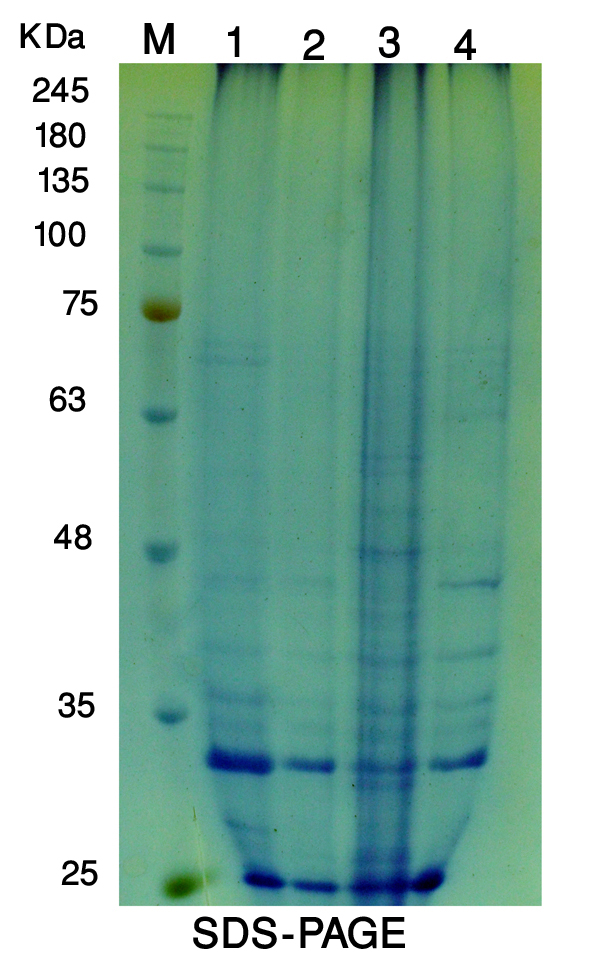 | Figure (8). Protein profile of Solanum nigrum callus control (1); callus exposure to 2mg/L AgNPs (2); 4mg/L (3) and 8mg/L (4) using SDS-PAGE technique. Marker (M) |
|
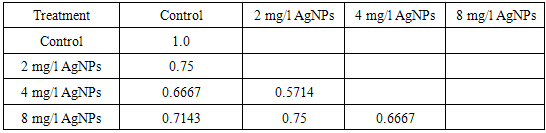
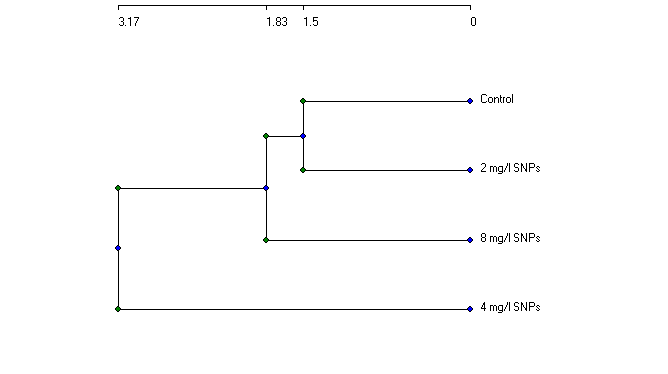 | Figure (9). Relationships between callus control (no AgNPs) and callus follwoed AgNPs at different concentrations (2, 4, 8 mg/l) based on protein profile |
|
3.5. Detection of Genetic Stability at DNA Level Using RAPD (PCR) Molecular Marker
- Random Amplified Polymorphic DNA (RAPD) marker was employed to detect level of genetic stability of callus of Solanum nigrum control (Lane 1) and callus treatment with 4.0mg/l AgNPs (lane 2). Six RAPD primers (OP-m01, OP-C15, OP-C12, OP-C04, OP-E15 and OP-B11) were selected according to their ability to detect distinct and clearly amplified products.
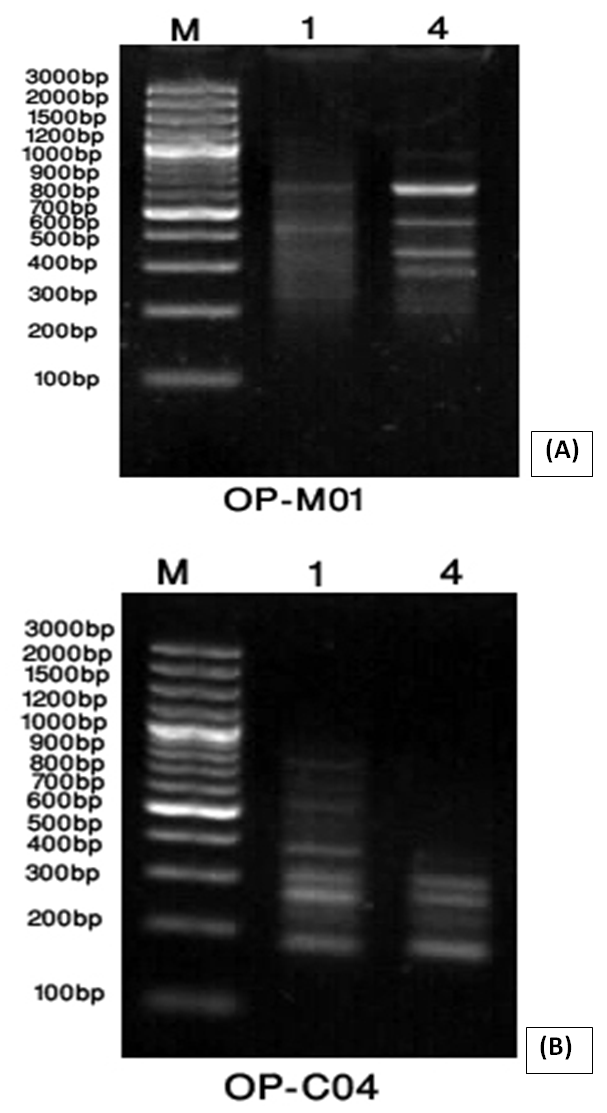 | Figure (9). A band pattern produced by primer OP-MO1 (A) and primer OP-CO4 (B) for callus control (1) and callus exposure to 4mg/L AgNPs (4). Line (M) represents DNA marker |
|
|
4. Conclusions
- Nanomaterials are considered to be one of the most important inventions of modern science [37]. In the present study, silver nanoparticles have been synthesized using safe, cost-effective, sustainable and environment friendly processes method (mixing 4 mL camel milk with 96 mL of 1 mM silver nitrate solution as previously mentioned). Their exceptionality is based on their constant physical properties, which are strictly dependent on their size, which varies from 1 to 100 nm. In addition, the interesting range of their properties is due to the large surface area that may be modified or functionalised for different biological applications [38]. These modifications can lead to higher water solubility or some targeting. Nanomaterials, especially nanoparticles, can be linked with different biologically active molecules, which can exactly direct them to specific sites within biomolecules including proteins and nucleic acids, sub-cellular, cellular, tissue and body structures [39]. Based on the results obtained in the present study, it can be concluded that uptake of metal nanoparticles (AgNPs) in vitro is connected with changes in Solanum nigrum callus morphology and anatomy, especially of biomass and deforming cell shape. Cell wall represents the main site of the interaction between nanoparticles and plant cell. This fact is probably connected with the effects on protein and nucleic acids metabolism that is clear with the variation observed in genetic stability of callus control (no AgNPs) and callus exposure to AgNPs.
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML