-
Paper Information
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
Nanoscience and Nanotechnology
p-ISSN: 2163-257X e-ISSN: 2163-2588
2015; 5(1): 14-21
doi:10.5923/j.nn.20150501.03
Biosynthesis of Silver Nanoparticles by Aspergillus oryzae (MTCC No. 1846) and Its Characterizations
Probin Phanjom, Giasuddin Ahmed
Department of Biotechnology Gauhati University, Guwahati, India
Correspondence to: Giasuddin Ahmed, Department of Biotechnology Gauhati University, Guwahati, India.
| Email: |  |
Copyright © 2015 Scientific & Academic Publishing. All Rights Reserved.
Biosynthesis of silver nanopartcles by Aspergillus oryzae (MTCC No. 1846) was demonstrated using both the fungal cell filtrate and the fungal biomass. In the present study, the fungal cell filtrate was used for extracellular biosynthesis of silver nanoparticles (AgNPs) from silver nitrate (AgNO3) solution. When the aqueous solution of 1mM AgNO3 was exposed to fungal cell filtrate, the aqueous silver (Ag+) ions were reduced to form extremely stable AgNPs in the solution The fungal biomass accumulated AgNPs on its surface, inside the cytoplasmic membrane and as well as within the cytoplasm in 72 hours (h) when challenged with 1mM AgNO3 solution. The formation of silver nanoparticles was characterized by UV–vis spectrophotometry and transmission electron microscope (TEM). X-ray diffractometer (XRD) spectrum of the nanoparticles confirmed the formation of metallic silver. The functional group of protein molecule surrounding the silver nanoparticles was identified using Fourier transform infrared spectroscopy (FTIR). The biosynthesized silver nanoparticles has zeta potential of -14.3 mV. The possible role of the enzyme nitrate reductase in the reducing process was also investigated. TEM studies showed the size of the silver nanoparticles to be in the range of 6nm to 26 nm.
Keywords: Silver nanoparticles, Aspergillus oryzae, TEM, SEM, XRD, FTIR, Zeta potential, Nitrate reductase
Cite this paper: Probin Phanjom, Giasuddin Ahmed, Biosynthesis of Silver Nanoparticles by Aspergillus oryzae (MTCC No. 1846) and Its Characterizations, Nanoscience and Nanotechnology, Vol. 5 No. 1, 2015, pp. 14-21. doi: 10.5923/j.nn.20150501.03.
Article Outline
1. Introduction
- Nanoparticles are usually referred to as particles with a maximum size of 100 nm. Application of nanoscale materials and structures usually ranges from 1-100 nm. Metal nanoparticles have a high specific surface area as well as high fraction of surface atoms. Because of their unique physicochemical characteristics such as size, distribution and morphology, they have been studied extensively for catalytic activity, optical properties, electronic properties, antibacterial properties, magnetic properties, biolabelling and treatment of cancer etc. [1–9]. Physical and chemical methods for nanoparticles synthesis are expensive and involve the production of toxic by products which are environmentally not safe methods. Depending on biological system as an alternative method for synthesizing nanoparticle and using microbes as a tool for synthesis of new functional nanomaterials has gained much interest in recent times [10, 11]. Nanoparticles synthesized using biological systems are also referred to as biogenic nanoparticles and the synthesis is classed as “green synthesis” or “green chemistry”. Many organisms synthesize AgNPs extra-cellularly, among which Fusarium oxysporum [12], Bacillus licheniformis [13], Aspergillus fumigatus [14], Klebsiella pneumoniae, [15], Bacillus subtilis [16], Escherichia coli [17], Aspergillus niger [18] and Aspergillus flavus [19] have been reported extensively. A novel green approach based on infrared ultra fast laser ablation to generate nanoparticles and bioconjugated nanoparticles has been reported recently [20, 21]. The filamentous fungi possess some distinctive advantages over bacteria viz like ease of handling, can be cultured on a large scale, high tolerance towards metals, as well as intracellular metal uptake capabilities [22]. Molecules performing dual nature such as capping and reducing are preferred since reaction can occur in one step without the use of any external reducing agents [23-25]. In the present study, a green approach to synthesize nanoparticles by using fungal cell filtrate of Aspergillus oryzae (MTCC No. 1846) without the use of chemical agents has been reported. The possible proteins present in the fungal cell filtrate serving as reducing and capping agent is also investigated.
2. Materials and Methods
2.1. Culture Maintenance
- The fungal strain Aspergillus oryzae (MTCC No. 1846) was obtained from Institute of Microbial Technology (IMTECH), Chandigarh, India. The strain Aspergillus oryzae was maintained at 25°C on czapek dox agar (CDA).
2.2. Biomass Production
- The fungal biomass was prepared by growing it aerobically in liquid broth of czapek dox (g/l) using Erlenmeyer flasks of 250 ml capacity containing 100ml of the media. The fungal strain was inoculated in the autoclaved media under sterilized and static conditions. The inoculum was allowed to grow for 5–7 days at 25°C. The medium was autoclaved at 15 psi for 30 min and cooled to room temperature before use. The pH of the medium was 5.6.
2.3. Synthesis of Silver Nanoparticles Using Cell Filtrate
- The fungus was grown aerobically in liquid broth of czapek dox (g/l). The culture flask was incubated at 25ºC and the biomass was harvested after 5 days of growth followed by extensive washing with sterile double distilled water to remove any medium components. Typically 10 g (wet weight) was brought into contact with 200mL sterile double distilled water for 5 days at 27ºC in an Erlenmeyer flask. After incubation, the fungal cell filtrate was obtained by passing it through Whatman filter paper No.1. After filtration, the observed pH of the cell filtrate was found to be 7.2. To the cell filtrate taken in 100ml capacity Erlenmeyer flask, 1mM AgNO3 was added and the reaction was carried out in dark condition at 27ºC. Simultaneously, a positive control of cell filtrate and a negative control containing only 1mM AgNO3 solution were maintained under same conditions. Sample of 3ml was withdrawn at fixed interval and the absorbance was taken.
2.4. Intracellular Synthesis of Silver Nanoparticles
- The fungus was grown aerobically in liquid broth of czapek dox (g/l). The culture flask was incubated at 27ºC and the biomass was harvested after 5 days of growth followed by extensive washing with sterile double distilled water to remove any medium components. Washed mycelium (4 g fresh wet weight) was challenged with 100 mL of 1mM AgNO3 solution (prepared in deionized water) of neutral pH and incubated in dark condition at 27ºC for 72 h. Simultaneously, a positive control of the fungal biomass incubated with deionized water and a negative control containing only 1mM AgNO3 solution were maintained under same conditions followed by visual monitoring.
2.5. Characterization of Synthesized Silver Nanoparticles
- The localized surface plasmon resonance of silver nanoparticles was characterized by using UV–Vis spectrophotometer (PC Based Double Beam Spectrophotometer, 2202, Systronic). The size and morphology of silver nanoparticles was characterized by TEM (JEOL JEM 100CX II) and the deposition of silver nanoparticles on the fungal surface was confirmed by SEM (JEOL JSM-6360). The presence of elemental silver was confirmed through X-ray diffraction (XRD) analysis (D8 ADVANCE, BRUKER). The interaction between protein–silver nanoparticles was analyzed by Fourier transforms infrared spectroscopy (FTIR, BRUKER Vector 22 Spectrophotometer) and the zeta potential was measured by Zeta sizer Nano-ZS90.
2.6. Nitrate Reductase Assay
- The enzyme-nitrate reductase in fungal filtrate was assayed according to the procedure followed by Harley [26]. Five milliliter aliquot of 5-day fungal filtrate was mixed with 5mL of assay medium (30mM KNO3 and 5% propanol in 0.1M phosphate buffer of pH 7.5) and incubated in the dark for 60 min. After incubation, nitrites formed in the assay mixture were estimated by adding 2.5 mL of sulphanilamide and NEED (N-(1-naphthyl) ethylene diamine dihydrochloride) solutions to it. The pink color developed was measured using UV–vis spectrophotometer. The enzyme activity was finally expressed in terms of n moles of nitrite/mL/h.
2.7. Determination of the Tryptophan / Tyrosine Residues
- Presence of tryptophan/tyrosine residues in proteins released by the fungal culture was analyzed spectrophotometrically by recording of the absorbance in the 250 – 300 nm wavelength regions according to the procedure followed by Saifuddin [27]. The recording of the absorbance were carried out by using UV–Vis spectrophotometer (PC Based Double Beam Spectrophotometer, 2202, Systronic).
3. Result and Discussion
3.1. Synthesis of Silver Nanoparticles Using Fungal Filtrate
- The Aspergillus oryzae cell filtrate prepared was pale yellow in colour and the pH was found to be 7.2. The fungal cell filtrate exhibited a gradual change in colour towards brown when it was incubated with 1mM AgNO3 in dark at 27ºC within 12 h of reactions which further darkened with increased incubation period time as shown Figure 1. This may be mainly because the reducing power of the responsible protein may acts as the reducing agent. The generation of dark brown colour is due to the surface plasmon resonance (SPR) exhibited by the nanoparticles.
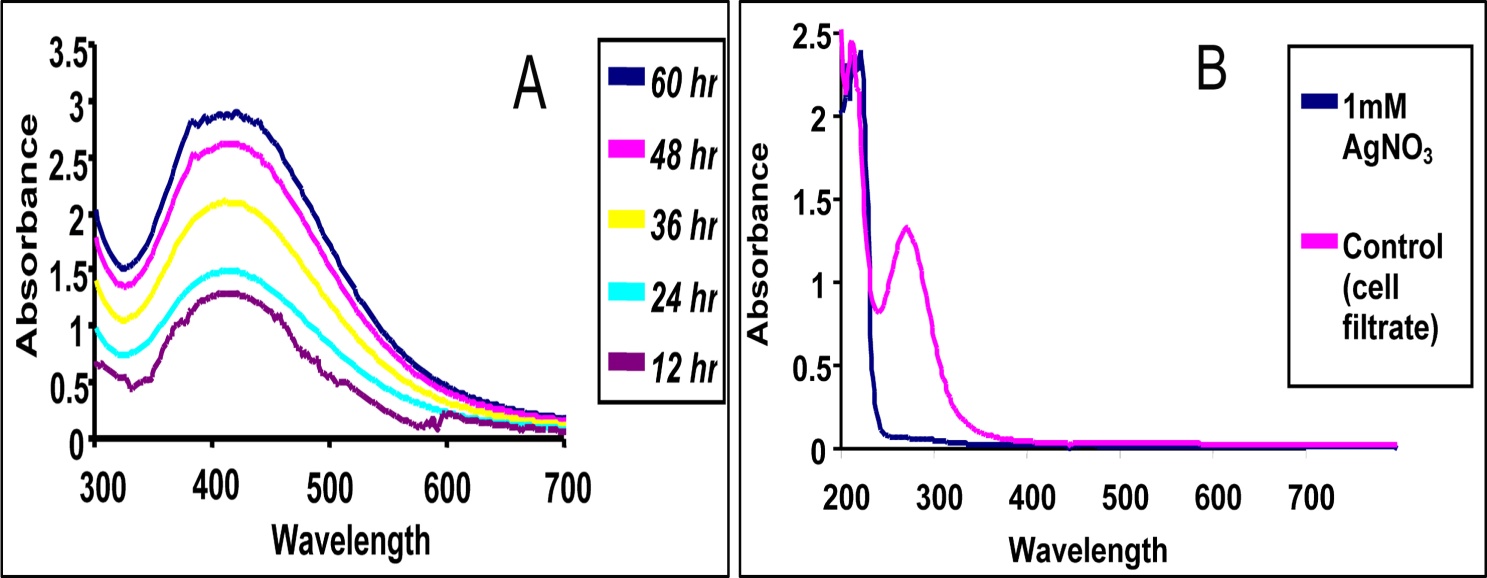 | Figure 2. UV–vis absorption spectrum of (A) Silver nanoparticles taken at different time interval and (B) 1mM AgNO3 and Control (Fungal Cell filtrate) |
3.2. Intracellular Synthesis of Silver Nanoparticles
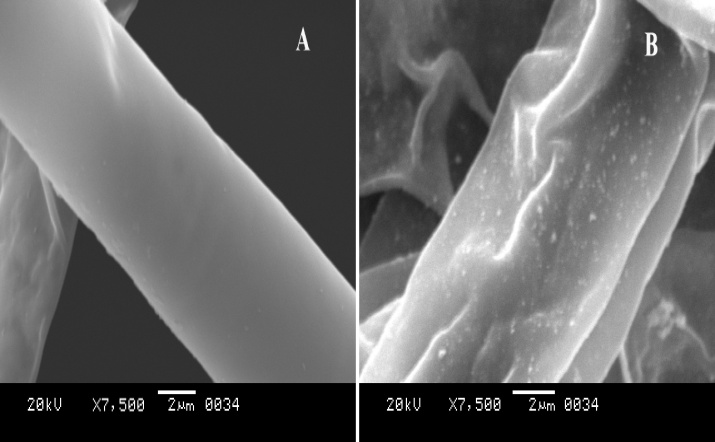
- When the fungal biomass was incubated for 72h at 27ºC with deionized water (positive control), it retained its original colour, while the fungal biomass challenged with 1mM AgNO3 solution turned brown after 72h, which further became dark brown after 7 days due to the deposition of silver nanoparticles. This colour is primarily due to the surface plasmon resonance of deposited silver nanoparticles as shown in Figure 3.
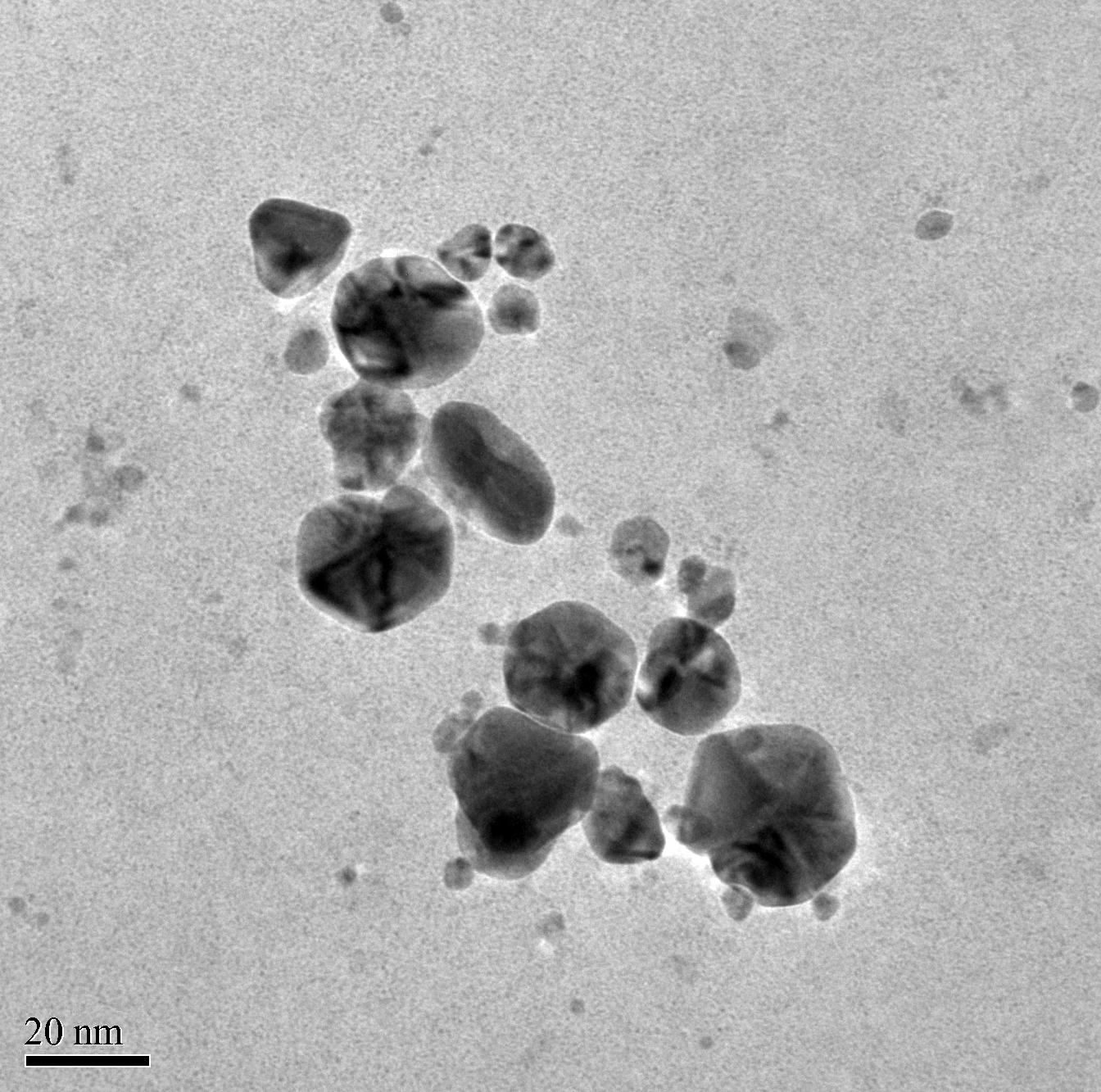
3.3. Transmission Electron Microscope and Scanning Electron Microscope Analysis
- Silver nanoparticles synthesized by the fungal cell filtrate was analysed by TEM as shown in the Figure 4. The morphology of the monodisperse silver nanoparticles is almost spherical. The particles size ranges from 6 nm to 26 nm having an average size of around 16.7 nm as shown in the Figure 5.
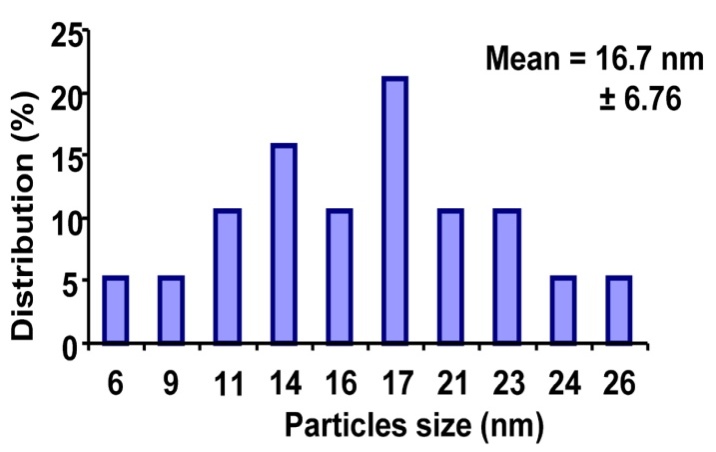 | Figure 5. Histogram of silver nanoparticles distribution |
3.4. X-ray Diffraction Analysis
- X-ray diffraction was carried out to confirm the crystalline nature of the particles, and the XRD pattern obtained is shown in Figure 8. The XRD pattern showed four intense representative XRD patterns of silver nanoparticles formed after reaction of fungal cell filtrate with 1mM AgNO3 aqueous solution for 24h at 27ºC. The XRD pattern showed four intense peaks in the whole spectrum of 2 theta values ranging from 35 to 80. The 2 theta peak values of around 38.10◦, 44.47◦, 64.63◦ and 77.44◦, were observed. A comparison of the obtained XRD spectrum with the standard confirmed that the silver particles formed in the experiments were in the form of nanocrystals, corresponding to [111], [200], [220] and [311], respectively, for silver.
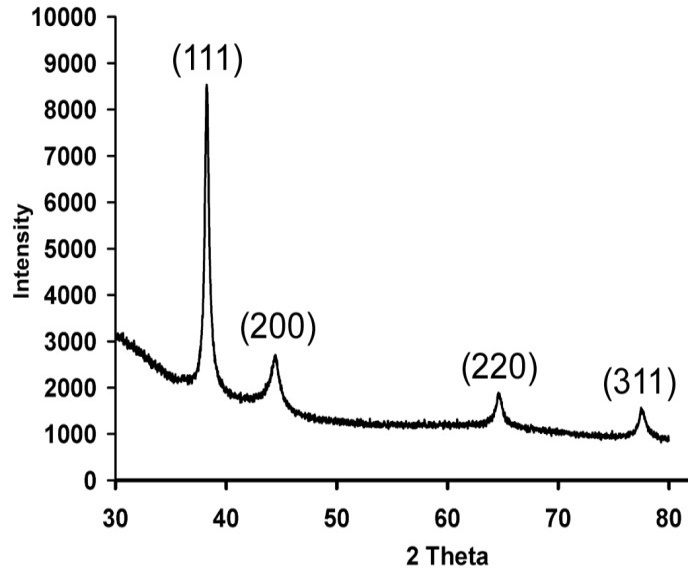 | Figure 8. XRD pattern of silver nanoparticles |
3.5. Detection of the Phenylalanine / Tryptophan / Tyrosine Residues
- The Figure 9 shows the UV-Vis spectrum in low wavelength region recorded for the fungal cell filtrate. The presence of the absorption band around 260 nm is attributed to the aromatic amino acids of proteins. There were two absorbance peaks found in the UV range corresponding to 210 and 260 nm. While the peak at 210 nm may be due to absorption by amide bond, the other peak at 260 nm may be attributed to the phenylalanine along with tyrosine and tryptophan residues present in the protein. This indicates secretion of some protein components into the medium by the fungal biomass which might play an important role in the reduction of the metal ions in the form of nanoparticles. Consequently the proteins may also bind to the nanoparticles and enhance the stability.
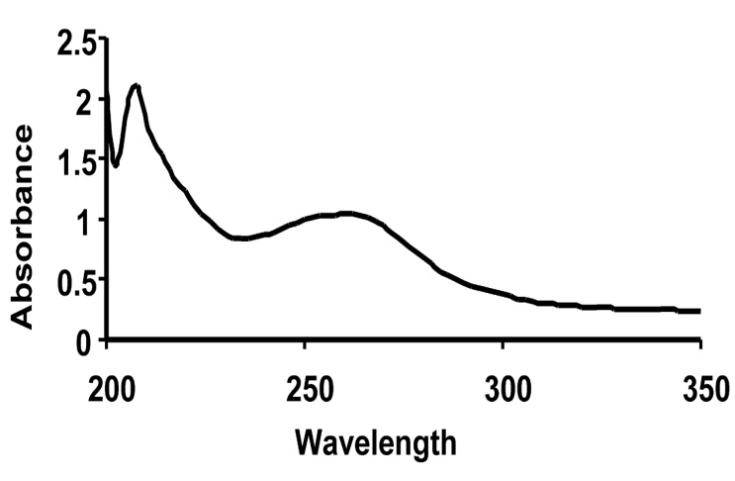 | Figure 9. UV-Vis spectrum in low wavelength region recorded for the fungal cell filtrate. There were two absorbance peaks found in the UV range corresponding to 210 nm and 260 nm |
3.6. FTIR Analysis
- The FTIR spectrum of biosynthesized silver nanoparticles showed eight distinct peaks, 1116, 1457, 1629, 2361, 2853, 2923 and 3421 cm−1 as shown in the Figure 10. The peak at 3421cm−1 and 2923 cm−1 refers to NH stretch vibration of primary and secondary amides of protein. The peak at 2853 cm−1 refers to C-H symmetrical stretch vibration of alkanes. The peak at 2361cm−1 refers to primary amine group of protein. The peak at 1629 cm−1 refers to carbonyl stretch, which is assigned to the amide I bond of protein. The peaks at 1457 refer to amino and amino-methyl stretching groups of protein. The peaks at 1116 cm−1 refer to C-O stretching vibrations mode. The carbonyl groups of the amino acid residues and the peptides have strong ability to bind to the silver [30]. It is also reported that the proteins can bind to nanoparticles either through free amine or cysteine groups in proteins [31, 32]. The proteins present over the silver nanoparticle surface may acts as capping agent for stabilization.
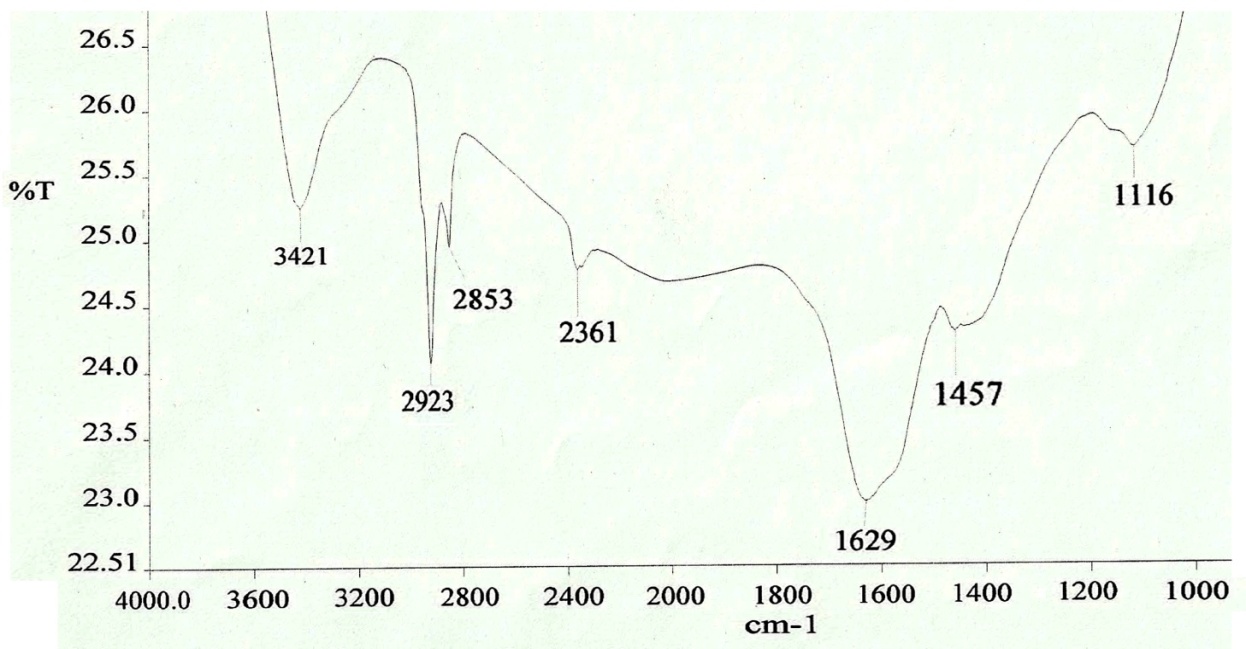 | Figure 10. FTIR spectrum of silver nanoparticles showed eight distinct peaks at FTIR spectrum of silver nanoparticles showed seven distinct peaks at 1116, 1457,1629,2361,2853,2923 and 3421 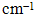 |
3.7. Zeta Potential
- The silver nanoparticles synthesized by the fungal cell filtrate were not in direct contact even within the aggregates, indicating stabilization of the nanoparticles, which was confirmed by measuring its zeta potential. The zeta potential indicates the degree of repulsion between adjacent, similarly charged particles in dispersion and its value can be related to the stability of colloidal dispersions. The zeta potential value of the biosynthesized silver nanoparticles was -14.3mV, indicating the presence of repulsion among the synthesized nano particles as shown in the Figure 11.
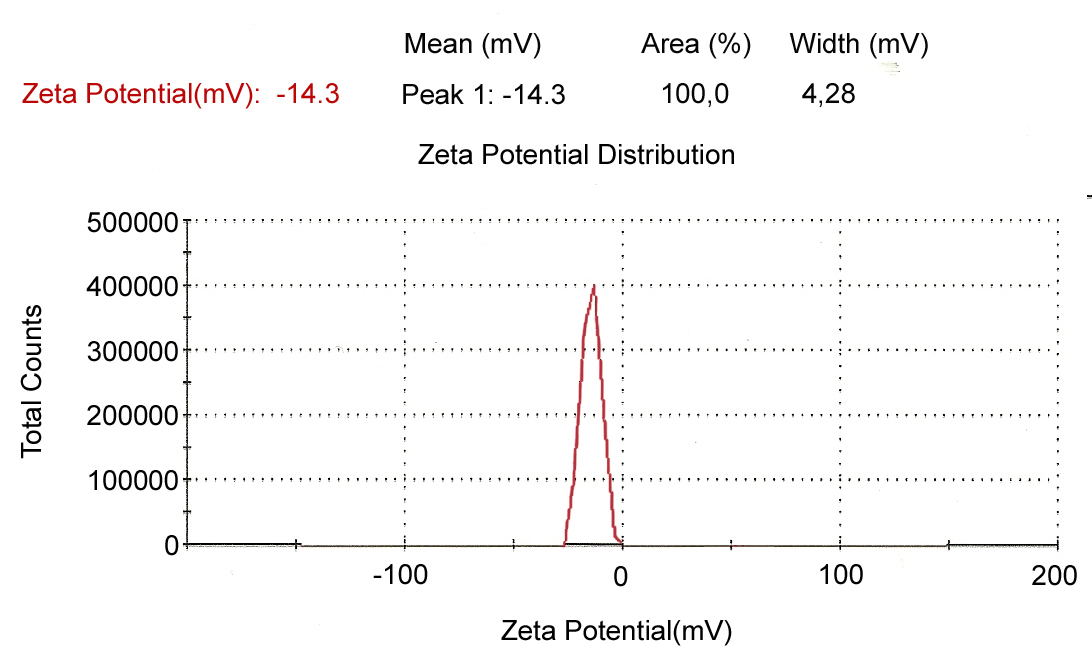 | Figure 11. Determination of surface charge of biosynthesized silver nanoparticles by zeta potential |
3.8. Nitrate Reductase Assay
- The extra cellular proteins secreted by the fungus may be responsible for the reduction of Ag+ to Ag0. Hence, the role of reductases in the fungal filtrate was investigated by nitrate reductase assay following the procedure given by Harvey. Fungal filtrate exhibited nitrate reductase activity in the present study. Duran et.al. [33] reported two possible mechanisms for the formation of silver nanoparticles by Fusarium oxysporum; one is through nitrate reductases and the other by shuttle quinine process. Similarly it is also reported that NADH-dependant nitrate reductase is the main enzyme responsible for the reduction of silver ions to silver in Fusarium oxysporum [34] and Bacillus licheniformis [35]. Therefore, nitrate reductase present in the fungal filtrate is implicated in the formation of silver nanoparticles.
4. Conclusions
- The studies above demonstrated extracellular synthesis of protein stabilized silver nanoparticles by Aspergillus oryzae without the use of any surfactants, protecting agents or linking agents under aqueous and ambient conditions. Intracellular deposition of silver nanoparticles was also observed within the mycelia. The coordination behaviors between amino groups in fungus and Ag+ ions are responsible for the stabilization of the silver nanoparticles. An extracellular enzyme-nitrate reductase appears to be involved in the synthesis of silver nanoparticles by Aspergillus oryzae, however more investigation is however required to further elucidate this aspect of reaction mechanism. The silver nanoparticles synthesized were almost spherical with sizes ranging from 6 nm to 26 nm. The silver nanoparticles were stable and the zeta potential was found to be -14.3mV.
ACKNOWLEDGEMENTS
- The authors are grateful to (SAIF) North Eastern Hill University, Shillong, India for TEM and SEM facilities, Department of Chemistry, North Eastern Hill University, Shillong, India for FTIR analysis and Institute of Advanced Study in Science and Technology (IASST), Guwahati, India for XRD analysis and zeta potential measurement. The authors would also like to thank the Department of Biotechnology, Gauhati University, Guwahati, Assam. India.
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML