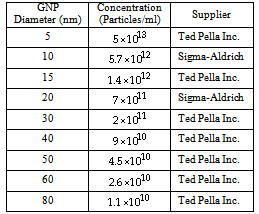
| [1] | Sperling R. A., Gil P. R., Zhang F., Zanella M., Parak W., “Biological applications of gold nanoparticles”, J., Chem. Soc. Rev., vol. 37, pp. 1896-1908, 2008. |
| [2] | Weibo C., Ting G., Hao H., Jiangtao S., “Applications of gold nanoparticles in cancer nanotechnology”, Nanotechnology Science Applications, vol. 1, pp. 17-32, 2008. |
| [3] | Devika B.Ch.,Arezou A.G., Warren C.W., “Determining the Size and Shape Dependence of Nanoparticle-Uptake into Mammalian Cells”, Nano Letters, vol. 6, pp. 662-668, 2006. |
| [4] | John M.T., Pure & Appl. Chem., 60, pp. 1517-1528, 1988. |
| [5] | Klaus, E.; Marion, L.; Martin, M., Matthias, P.; Helmut, S., Method for manufacturing substrates with transparent and color coating stable at high temperature and in the presence of ultraviolet rays, U.S. Patent, 6156388, Dec. 5, 2000. |
| [6] | Bing T., Yiying W., “Dye-Sensitized Solar Cells Based on Anatase TiO2 Nanoparticle/Nanowire Composites”, J. Phys. Chem.. B, 110, pp. 15932-15938, 2006. |
| [7] | Frank C., “Nanoengineering of Particle Surfaces”, Adv. Mater., 13, pp. 11-22, 2001 |
| [8] | Marie Ch., Didier A., “Gold Nanoparticles: Assembly, Supramolecular Chemistry, Quantum-Size-Related Properties, and Applications toward Biology, Catalysis, and Nanotechnology”, Chem. Rev., 104, pp. 293-346, 2004. |
| [9] | Sang B. L., “Focus on nanoparticles for cancer diagnosis and therapeutics”, Nanomedicine, vol. 2, no. 5, pp. 647-648, 2007. |
| [10] | Hainfeld J.F., Slatkin D.N., Focella T.M., Smilowitz H.M., “Gold nanoparticles: a new X-ray contrast agent”, The Britsh Journal of Radiology, 79, pp. 248-253, 2006. |
| [11] | Xiaohua H., Ivan H., Wei Q., Mostafa A., “Cancer Cells Assemble and Align Gold Nanorods Conjugated to Antibodies to Produce Highly Enhanced, Sharp, and Polarized Surface Raman Spectra: A Potential Cancer Diagnostic Marker”, Nano Letters, vol. 7, no. 6, pp. 1591-1597, 2007. |
| [12] | Mauro F., “Cancer nanotechnology: opportunities and challenges”, Nature Reviews Cancer, vol. 5, no. 3, pp. 161-171, 2005. |
| [13] | Tian Z.Q., “Surface-enhanced Raman spectroscopy: advancements and applications”, J. Raman Spectrosc., vol. 36, pp. 466-470, 2005. |
| [14] | Kneipp K.., Haka A.S., Kneipp H., Badizadegan K., Yoshizawa N., Boone C., Shafer P., Motz J., Dasari R., Feld M., “Surface-Enhanced Raman Spectroscopy in Single Living Cells Using Gold Nanoparticles”, Applied Spectroscopy, 2002, vol. 56, no. 2, pp. 150-154. |
| [15] | Hope T.B., Christopher B.C., I-Hsien, C., James P., James E.H., Melodie E.B., Joseph B. J., Theresa A.G., Gerard L.C., “Application of Surface Enhanced Raman Spectroscopy for Detection of Beta Amyloid Using Nanoshells”, Plasmonics, vol. 2, pp. 55-64, 2007. |
| [16] | Daniel M. K., Brian G. P., Orlin D.V., “Controlled assembly of SERS substrates templated by colloidal crystal films”, J.Mater.Chem, vol. 16, pp. 1207-1211, 2006. |
| [17] | Wolfgang H., Nguyen T. K., Thanh J. A., David G. F., “Determination of Size and Concentration of Gold Nanoparticles from UV-Vis Spectra”, Anal. Chem., vol. 79, pp. 4215-4221, 2007. |
| [18] | Nikhil R.J., Latha G., Catherine J. M., “Seeding Growth for Size Control of 5−40 nm Diameter Gold Nanoparticles”, Langmuir, vol. 17, pp. 6782-6786, 2001. |
| [19] | Kenneth R.B., Daniel G.W., Michael J.N., “Seeding of Colloidal Au Nanoparticle Solutions. 2. Improved Control of Particle Size and Shape”, Chem. Mater., vol. 12, no. 2, pp. 306-313, 2000. |
| [20] | Jan J.G., “On the relationships between SVD, KLT and PCA”, pattern recognition, vol. 14, no. 1-6, pp. 375-381, 1981. |
| [21] | Salim C., Amrane H., Boualem S., “A new PCA-based method for data compression and enhancement of multi-frequency polarimetric SAR imagery”, J. Intelligent data analysis, 6, pp. 187-207, 2002. |
| [22] | Mia H., Sanne E., “Robust PCA and classification in biosciences”, Bioinformatics, vol. 20, no. 11, pp. 1728-1736, 2004. |
| [23] | Pichardo J.L., Frausto C., Barbosa O., Huerta, R., Gonzalez J.L., Ramírez C.A., Gutierrez G., Medina C., “Raman spectroscopy and multivariate analysis of serum samples from breast cancer patients”, Lasers Med. Sci., 22, pp. 229-236, 2007. |
| [24] | González J.L., Rodríguez J., Martínez J.C., Frausto, C., Jave L., Aguilar A., Vargas H., Martínez E., presented at Laser Florence 2009, International Congress Laser Medicine with Pre conference Courses, November 6-November 7, Springer, Florence 2009, 12(26): pp. 91-95. |
| [25] | Martínez J.C., González J.L., Frausto C., Miranda M.L., Soria C., Medina J., presented at Laser Florence 2008, International Congress Laser Medicine with Pre conference Courses, October 31-November 1, Springer, Florence 2008, vol. 23: pp. 99-103. |
| [26] | Chad A.L., Anita M.J., “Automated method for subtraction of fluorescence from biological Raman spectra”, Applied spectroscopy, vol. 57, no. 11, pp. 1363-1367, 2003. |
| [27] | Hans F.M., Win T. K., Onno E.N., Age K.S., “Performance Optimization of Spectroscopic Process Analyzers”, Anal.Chem., vol. 76, no. 9, pp. 2656-2663, 2004. |
| [28] | Jirasek A., Schulze G., Yu, M., Blades M., Turner R., “Accuracy and Precision of Manual Baseline Determination”, Applied Spectroscopy, vol. 58, no.12, pp. 1488-1499, 2004. |
| [29] | Aleix M.M., Avinash C. K., “PCA versus LDA”, IEEE transactions on pattern analysis and machine intelligence, vol. 23, no. 2, pp. 228-233, 2001. |
| [30] | Sharad M., Eamonn, K., Kaushik C., Michael P., “Locally adaptive dimensionality reduction for indexing large time series databases”, ACM transactions on database systems, 27, pp. 188-228, 2002. |
| [31] | Saurav D., Goutam N., Asish B., “Application of PCA-based hybrid Taguchi method for correlated multicriteria optimization of submerged arc weld: a case study”, Int. J. Adv. Manuf. Technol., vol. 45, pp. 276-286, 2009. |
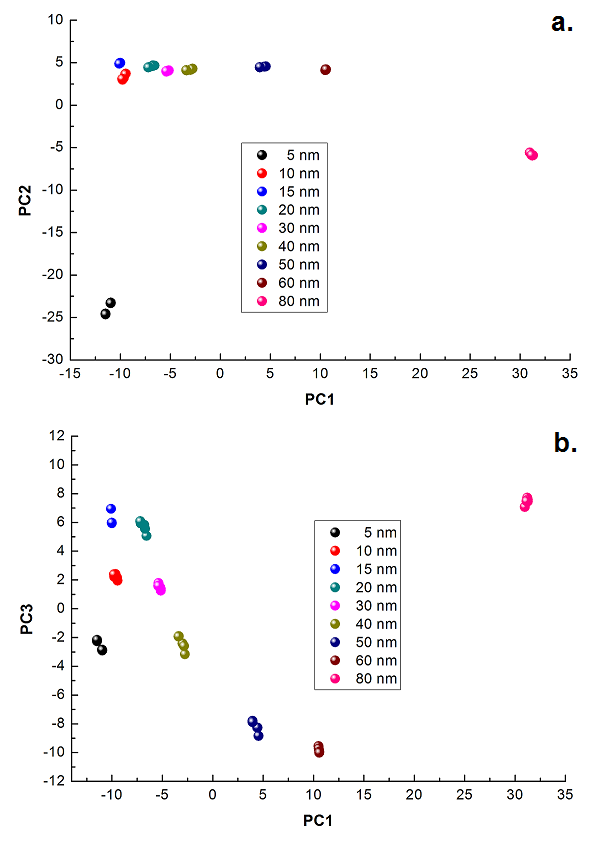
 )
)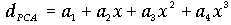
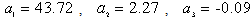 , and
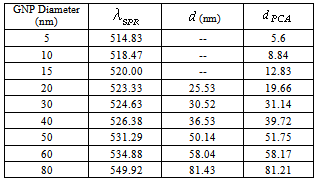
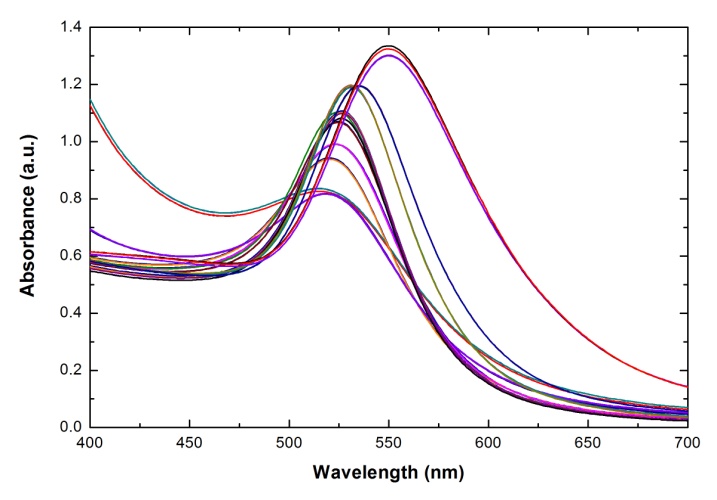
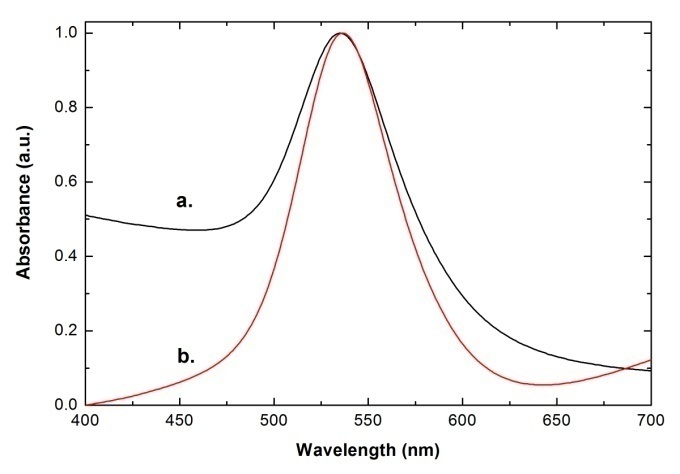
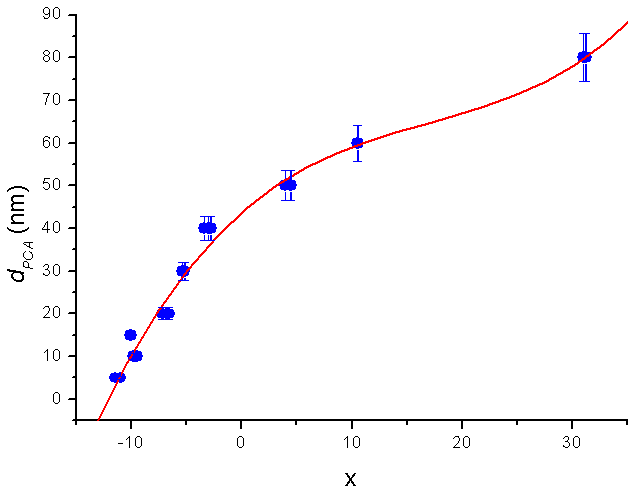
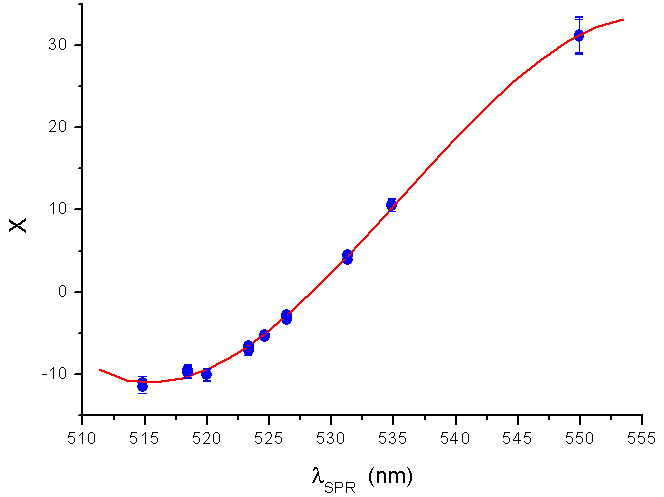
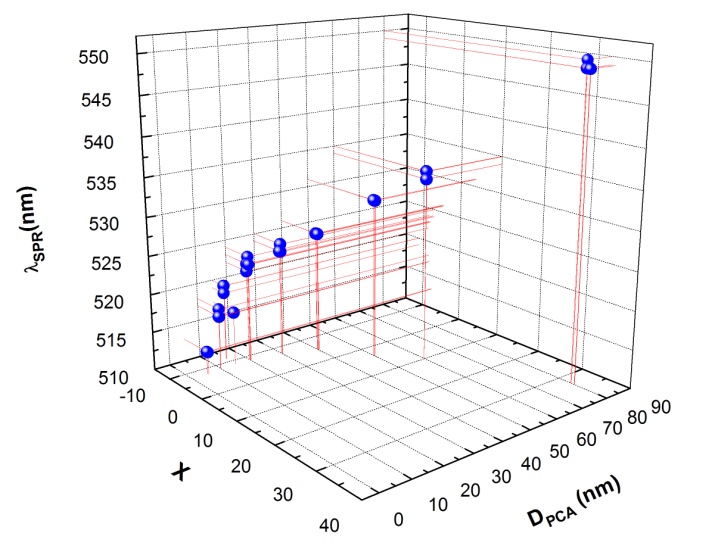
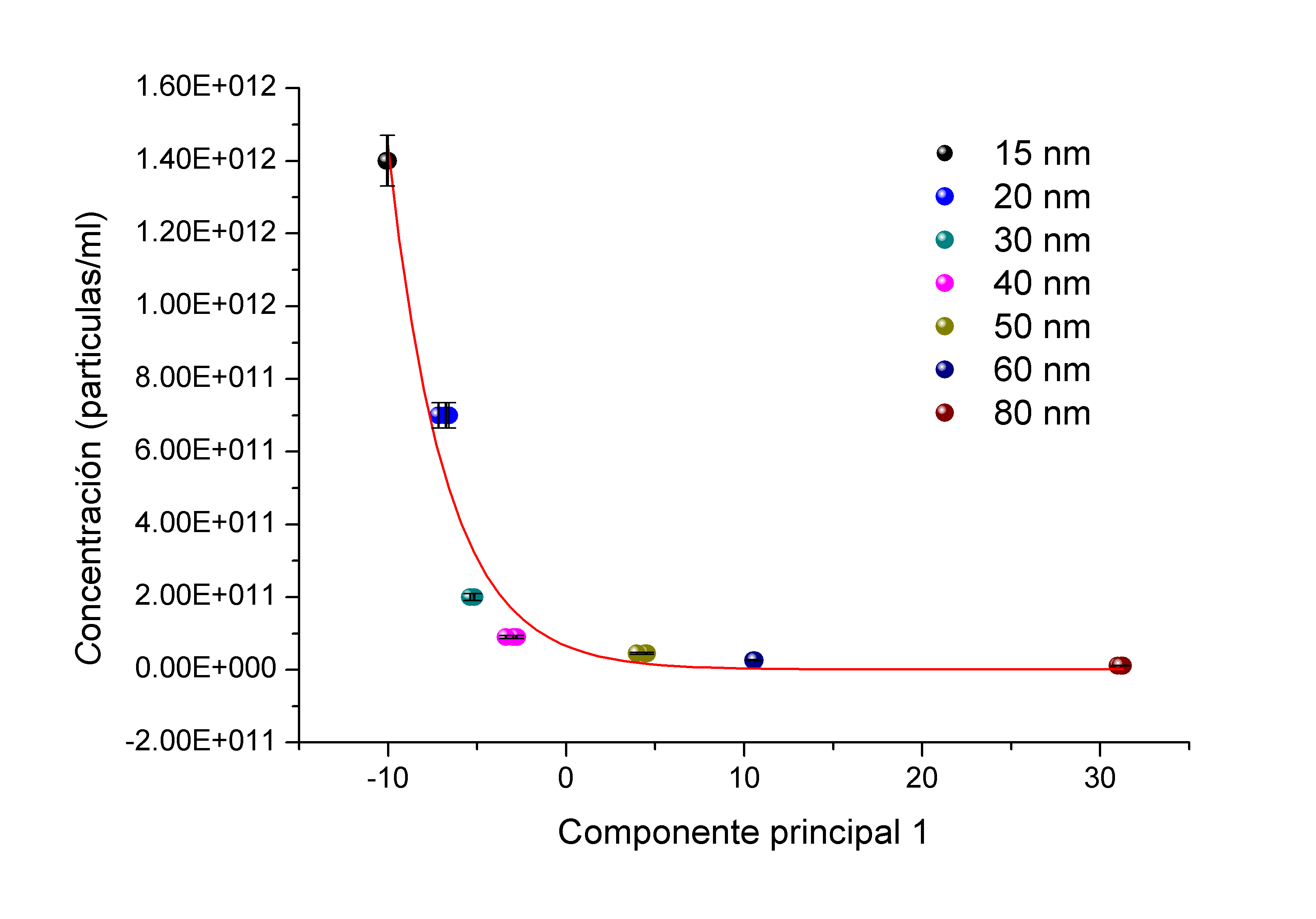
, and 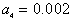 and the result of fitting to a polynomial of degree 3 between the first principal component and the diameter of the GNP.In the figure 5 is shown the value graph for the dominant component vs. the position of the Surface Plasmon Resonance peak in the spectrum of each suspension, it is found a systematic variation. This one can be fitted to a third order polynomial function for wave length in nm (
and the result of fitting to a polynomial of degree 3 between the first principal component and the diameter of the GNP.In the figure 5 is shown the value graph for the dominant component vs. the position of the Surface Plasmon Resonance peak in the spectrum of each suspension, it is found a systematic variation. This one can be fitted to a third order polynomial function for wave length in nm (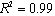 ).
).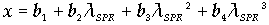
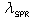 is the position of the SPR peak and the fit parameters,
is the position of the SPR peak and the fit parameters, 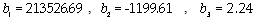 and
and 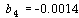 .
. 
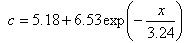
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-Text HTML
Full-Text HTML