-
Paper Information
- Next Paper
- Previous Paper
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
Marine Science
p-ISSN: 2163-2421 e-ISSN: 2163-243X
2012; 2(5): 57-65
doi: 10.5923/j.ms.20120205.04
Cryptic Species: A Mismatch between Genetics and Morphology in Millepora
Craig Tepper 1, Logan Squiers 2, Chuck Hay 1, Danielle Gorbach 1, Dana Friend 3, Bob Black 1, Ben Greenstein 3, Kevin Strychar 2
1Department of Biology, Cornell College, Mt. Vernon, IA 52314, USA
2Life Sciences Department, Texas A&M-Corpus Christi, Corpus Christi, TX 78412, USA
3Department of Geology, Cornell College, Mt. Vernon, IA 52314, USA
Correspondence to: Craig Tepper , Department of Biology, Cornell College, Mt. Vernon, IA 52314, USA.
| Email: |  |
Copyright © 2012 Scientific & Academic Publishing. All Rights Reserved.
Millepore morphology is highly variable and shows signs of phenotypic plasticity. Two species of Millepora are present around the islands of the Bahamas: one exhibiting a strong, blade-like structure, Millepora complanata, and the other having a delicate branch-like structure, Millepora alcicornis. The phylogenetic relationship of these corals has been under considerable debate for many years. The existence of a range of intermediate growth forms exhibiting characteristics of both recognized species has led to the re-examination of this species complex. Several methods were employed to examine the taxonomic relationship including ecological abundance surveys, morphological thin-section analysis, and sequencing of rDNA internal transcribed spacer (ITS) regions. Abundance surveys showed a demarcation of growth forms by depth at two sites but an intermingling of growth forms at a third site. Morphometric analysis resulted in discrimination between M. alcicornis, M. complanata and the intermediate growth forms. However, rDNA sequence differences revealed the presence of two distinct clades, each containing members of the two currently recognized species as well as intermediate growth forms. The sequence analysis suggests the presence of two, phenotypically plastic cryptic species. Although limited in scope, our results indicate that caution should be exercised when describing species based on morphology alone and that multiple characters, including genetic information, should be used when describing species relationships.
Keywords: Millepora, Phenotypic Plasticity, Morphometric, Cryptic Species, rDNA, ITS
Article Outline
1. Introduction
- The genus Millepora (family Milleporidae, classHydrozoa, phylum Cnidaria), commonly referred to as “fire-coral,” is an integral part of reef communities[1]. Fire corals serve as important framework builders, second only to scleractinian corals[2]. This framework is supported by an algal zooxanthellate symbiont, which aids in light-enhanced calcification[3]. Millepores are distributed worldwide in tropical seas and typically range in depth from less than 1m to approximately 40m[4]. The habitats they live in can range, depending mostly on growth form, from strong turbulent shallow waters to sheltered deeper waters[5]. Themorphology of the millepores is highly variable and is believed to show phenotypic plasticity[1,6].Phenotypic plasticity is believed to be molded by selection, such that certain phenotypes are better able to exploit a given environment[7]. Hence, a given species may exhibit different phenotypes depending on the environment. Plasticity has been described in many marine taxa including corals[8-9], sponges[10], fish[11], barnacles[12], and mollusks[13]. The presence and range of taxa shown to exhibit phenotypic plasticity must be taken into consideration when applying morphological characters to define species. The various growth forms of Millepora in the Caribbean (Figure 1) range from thinly encrusting sheets and delicate dendroid branches, M. alcicornis, to thicker, rigid bladed forms, M. complanata[1]. It is this variation in morphology that has led to constant controversy about Millepore classification. Debate about taxonomy is not restricted to the millepores; morphological plasticity has also been documented in many scleractinian corals[14].Since Millepora was first recognized by Linnaeus in 1758, many naturalists have worked on the millepores and their research has resulted in numerous and widely varied classification schemes[6]. When first described, millepores were primarily classified using morphological characters such as texture of the surface of the coral, size and shape of pores, stinging properties, and visual appearance[15]. Early investigators[16] found Millepora to be quite diverse and recognized 22 different species from the Caribbean. Later, Hickson[17] suggested that all recognized species were environmentally controlled growth forms of a single Millepora species. Boschma[4] recognized ten species of Millepora, three from the Atlantic Ocean and seven from the Indian and Pacific Oceans. Although 50 species of millepores have been described by Vernon[18], currently there are 17 recognized extant Millepora species; 11 from the Pacific and six from the Atlantic[19-20]. Wide variation in growth form of all species and a lack of diagnostic morphological characters presents serious problems for correct identification at the species level.
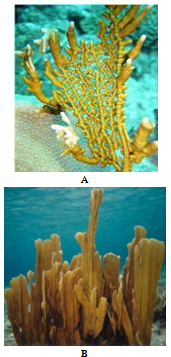 | Figure 1. Photographs depicting the typical growth forms of Millepora species found in the Bahamas. A Millepora alcicornis B Millepora complanata |
 | Figure 2. Photographs depicting examples of intermediate growth forms of Millepora found around San Salvador Island, Bahamas |
2. Materials and Methods
2.1. Collection Sites
- Millepores used in this study were collected from reefs surrounding the island of San Salvador, Bahamas (Figure 3). San Salvador is located on the eastern edge of the Bahamas Island chain and is characterized by its karst and hypersaline lakes. Patch reefs included for collection were Lindsay Reef (24°00’32”N, 74°31’59”W), Rocky Point Reef (24°06’25”N, 74°31’17”W), and French Bay (23°56’59”N, 74°32’50”W) (Figure 3). Lindsay Reef and Rocky Point Reef are shallow reefs with maximum depths of approximately 5m. French Bay has both shallow (1-5m) and deep (5-10m) patch reefs.
2.2. Ecological Abundance Surveys
- Twenty meter line transects were laid down at random points at both French Bay and Lindsay Reef. Transects 1-3m in depth were surveyed by snorkeling, and transects deeper than 3m were conducted using SCUBA. All Millepora colonies within one meter of either side of an outstretched tape measure were recorded giving a total area surveyed for each transect of 40m2. Only colonies with the classic finely branched morphology were considered to be M. alcicornis and only bladed colonies with a complete absence of branches were considered to be M. complanata. Millepora colonies not meeting these specifications were recorded as intermediate growth forms.
2.3. Sample Collections
- Coral samples were randomly collected from each of the aforementioned reefs by removing a small piece, approxi-. mately 4 sq. cm in size. Samples of M. alcicornis, M. complanata, and the intermediate morphology were transported from the collection sites to the Gerace Research Centre (San Salvador, Bahamas) in buckets containing seawater and held in a flow-through seawater tank for no more than two days until coral DNA was isolated.
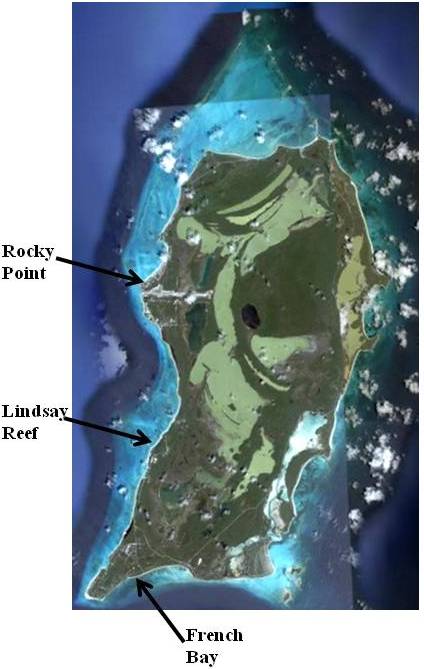 | Figure 3. Satellite image San Salvador, Bahamas showing the three collection sites[27] |
2.4. Morphometric Analysis
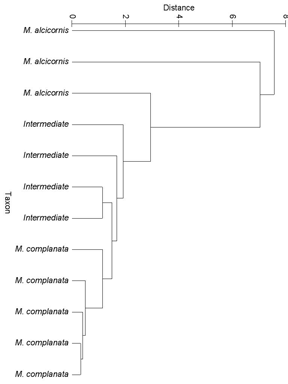
- Twelve specimens of Millepora were subjected to morphometric analysis following the method outlined by Amaral et al.[28]. Specimens included five individuals assigned to M. complanata, three individuals of M. alcicornis and four individuals that exhibited intermediate growth forms.Specimens were cut and mounted onto standardmicroscope slides, ground to standard thin sections (30µ) andanalyzed using a petrographic microscope. A series of 25 mm2 grids was superimposed on each thin section such that replicate, non-overlapping grids could be analyzed. Each grid was photographed using a SONY ExwaveHAD digital video camera and imported into Photoshop for analysis.A variety of features of the skeletal microstructure were quantified using the Analysis Tools module in Photoshop. These included diameter and surface area of all gastropores and dactylopores, distance between gastropores,dactylopores, and their density (# pores/grid). The number of dactylopores associated with each gastropore also was recorded. Variable numbers of individual gastropores (8-23) and dactylopores (27-129) were measured from each grid. Total number of grids measured was determined by the surface area of the hydrozoan present on each thin section. N = 3-5 grids per coral (8-129/grid, see above). Within-and between-grid values were averaged. Since no difference was observed using pooled vs. unpooled data, pooled data from each specimen are presented here. A Q-mode distance matrix was generated from the average values obtained from each specimen using the Euclidean Distance Coefficient. The matrix was subjected to Cluster Analysis (UPGMAA algorithm). An Analysis of Similarity (ANOSIM)[29] was performed to assess significance of the differences observed between samples (both procedures in Primer v. 6.15). The Q-mode dendrogram was compared to the results of molecular genetic analysis.
2.5. DNA Techniques
- Genomic DNA was isolated using a procedure modified from Rowan and Powers[30] and Lopez et al.,[31]. Coral tissue was removed by repeatedly blasting the skeleton with a 50cc syringe containing L buffer (100mM EDTA, 10mM Tris, pH 7.6). Coral tissue was centrifuged at 3500rpm for 10 minutes; the resulting pellet was washed in 10mL of L buffer and re-centrifuged. The tissue pellet was resuspended in 900µL of L buffer and macerated manually with a tissue homogenizer. The homogenate was centrifuged twice at 500rpm for 10 minutes in order to separate the coral tissue from the liberated zooxanthellae. Following the addition of 1% (w/v) SDS to the supernatant, the lysate was incubated at 65℃ for 30-60 minutes. Pro K (0.5 mg/mL) was added and the lysate was incubated at 37℃ for at least 6 hours. NaCl (0.8M) and CTAB (1% w/v) were added and the sample was incubated at 65℃ for 30 minutes. Nucleic acids were precipitated twice in 70% (v/v) ethanol and 3M sodium acetate (pH 5.2) and immediately centrifuged. Following resuspension of the pellet in dH2O, the DNA was briefly centrifuged and the supernatant was retained.ITS rDNA PCR amplification was performed using 100-300ng of template, 60pmol of the coral specific primer A18S (5’-GATCGAAC-GGTTTAGTGAGG-3’) and 60pmol of the universal primer ITS 4 (5’-TCCTCCGCTTA TTGATATGC-3’)[25], 10X Tfl PCR buffer (Promega, Madison, WI), 2.0mM MgSO4, 0.1mM dNTP and 1U of Tfl polymerase. The PCR profile was: 1 cycle of 94℃ for 2 minutes; 30 cycles of 94℃ for 1 minute, 55℃ for 2 minutes, and 72℃ for 3 minutes; and 1 cycle of 72℃ for 5 minutes[26]. Amplified PCR products were run on 1.2% (w/v) low melting agarose gels. Discreet, prominent bands were excised and purified using the Wizard SV Gel and PCR Clean-Up System (Promega). Purified PCR products were ligated into pGEM-T vectors following the manufacturer’s protocol (Promega) and transformed into competent DH5 E. coli host cells. Following blue-white selection, positive colonies were harvested and plasmid DNA was isolated using the Zyppy Plasmid Miniprep Kit (Zymo Research, Orange, CA). Plasmids containing ITS rDNA inserts were sequenced in both directions with 3.0 pmol of M13 forward and reverse primers. Reagents and reaction conditions for sequencing were as specified by the USB Thermo Sequenase Cycling Sequencing Kit (USB, Cleveland, OH). PCR products were run in 5.5% KBPlus Gel Matrix acrylamide using a LI-COR 4300 DNA Analyzer (LI-COR, Lincoln, NE). Sequence reaction products were analyzed using e-Seq V3.0 (LI-COR).
2.6. Phylogenetic Analysis
- Maximum likelihood trees were produced using MEGA 4.0.1. The sequence of Millepora exaesa[32] was used for the outgroup (GenBank, accession no. U65484) and 1000 bootstrap replicates were performed. Nucleotide percent substitution was also calculated from sequence data and compared within and between morphologies using MEGA 4[33] (The Biodesign Institute, Tempe, AZ).
3. Results
3.1. Abundance Surveys
3.2. Morphometric Analysis
- Morphometric data were able to discriminate between the two standard Milleporid taxa (M. complanata and M. alcicornis) as well as those specimens exhibiting an intermediate growth form (Figure 5). Specimens of M. alcicornis are “tacked on” to the cluster containing the intermediate growth forms at relatively low similarity values.
3.3. PCR Amplification of rDNA
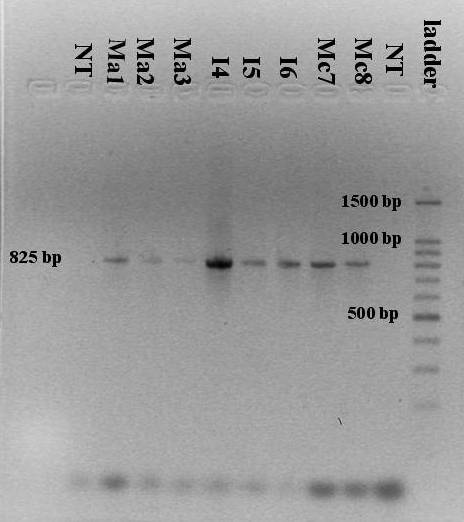
- Fragments of the ITS rDNA region (18S rDNA, ITS-1, 5.8S rDNA, ITS-2, and 28S rDNA) were amplified from bladed, branching and intermediate growth forms and a single PCR amplification product of approximately 825 base pairs was obtained from every sample (Figure 6).
3.4. DNA Sequence Analysis
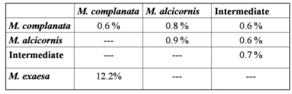
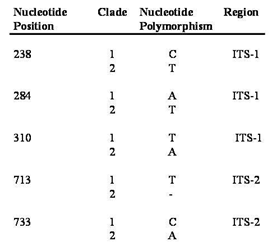
- A total of 36 samples were sequenced (17 of M. complanata, 12 of M. alcicornis, 7 intermediate growth forms) each yielding a sequence length of approximately 825 base pairs. The aligned sequences including gaps were 843 sites long and contained 778 conserved sites, 50 variable sites, and 10 parsimony informative sites.Genetic variation, as nucleotide percent substitution, both within and between morphologies of M. alcicornis, M. complanata and intermediate growth forms was relatively low ranging from 0.6% to 0.9% (Table 1). Nucleotide substitution rates were higher when M. complanata was compared to M. exaesa from the Red Sea[32].
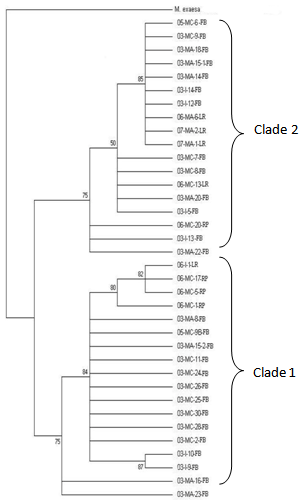 | Figure 7. Maximum likelihood tree showing bootstrap values for 36 samples of Millepora from the full range of phenotypes. 03, 05 or 06 denote the year the sample was collected (2003, 2005 or 2006). MA = Millepora alcicornis, MC = Millepora complanata, I = intermediate growth form. Numbers after the species designations represent the sample number. FB = French Bay, LR = Lindsay Reef and RP = Rocky Point. Bootstrap values are listed on the branchpoints. M. exaesa was used as the outgroup |
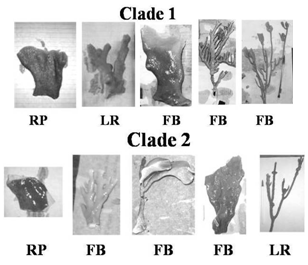 | Figure 8. Representatives of Millepora samples that are classified as clade 1 and 2. Collection sites are FB = French Bay reef, LR = Lindsay Reef, and RP = Rocky Point reef |
|
4. Discussion
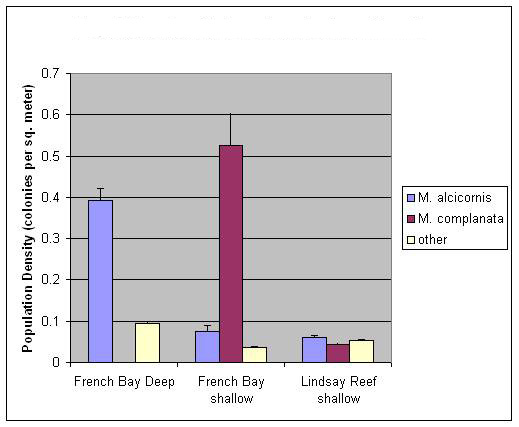
- The taxonomy of the millepores is currently based on morphological characters and does not take into account genetic differences that may be present[15]. Taken alone, our morphometric results corroborate earlier work that distinguishes between the millepores based on morphology. The fact that M. alcicornis was not as clearly differentiated by our cluster analysis may in part be the result of a smaller sample size of this species in thin section (two-dimensional planes obtained from branching hydrozoan colonies are quite small). A more interesting result is the contrast between the outcomes produced by genetic and morphometric data, which suggests that the standard, morphologically based, taxonomy may be incorrect; two species of Millepora may exist around San Salvador that cannot be distinguished based upon morphology alone. This hypothesis suggests that two phenotypically plastic cryptic species are present and appear to be reproductively isolated from one another.Both currently recognized Millepora species are morphologically plastic and their relative abundance was found to be different between study sites. Abundance surveys at French Bay suggest the two morphologies may be utilizing different habitats since one is predominantly found in shallower waters, while the other is found almost exclusively in deeper waters (Figure 4). At first glance, this data seems to support the current taxonomy, but when phenotypic plasticity is incorporated, the various growth forms present may simply be the result of the different environments in which they are found. Additionally, the occurrence of all growth forms in mutual proximity on Lindsay Reef supports Stearn and Riding’s[1] contention that morphological variation in the Millepores is not primarily a response of a single species to environmental differences. Results from abundance surveys at Lindsay Reef suggest that genetic differences may exist between the growth forms.Results from thin section analysis support the abundance data but suggests a different taxonomic relationship than the sequence data. Morphometric results indicate that pore size and arrangement can be used as a diagnostic tool to distinguish the two species of Millepora (Figure 5). A possible explanation for this discrepancy with the genetic data is again phenotypic plasticity. While the environment has been shown to alter macro-morphological characteristics via plasticity, it is not much of a stretch to suggest that the environment can alter micro-morphological characters, such as pore size, as well. While more work needs to be done in this area, it is our assertion that the environment plays a large role in determining the macro and micro morphologies of these corals. Results from the ITS rDNA sequence comparison of the two purported species of Millepora and the intermediate growth forms show that the three morphologies are very closely related (Table 1). However, the rDNA ITS region exhibits considerable divergence when compared to the reproductively isolated M. exaesa found in the Red Sea. Takabayashi et al.[24] have shown that the size of the PCR amplified ITS rDNA fragment may be used as a diagnostic marker to distinguish between closely related species. Meroz-Fine et al.[26] reported that two growth forms of M. dichotoma that were classified as a single species contained ITS rDNA regions that were quite different in size and sequence leading them to conclude the two growth forms were different species. Regardless of the growth form sequenced, all of our millepore samples had a fragment length of approximately 825 base pairs suggesting that the millepores may be one plastic species. However, the phylogenetic tree (Figure 7) generated from these sequences demonstrated the presence of two clades, each containing members of all three morphologies (Figure 8), which further suggests the current taxonomy of Millepora may not be accurate. The conservation of the SNP pattern (Table 2) suggests that these two clades are reproductively isolated and the intermediate growth forms are not a result of extensive hybridization.However, basing species-level phylogeneticreconstructions on ITS regions is sometimes problematic due to intragenomic sequence variation in the rDNA tandem repeats[34]. Variant rDNA copies can arise spontaneously in a single generation from point mutations. LaJeunesse and Pinzon[35] maintain that the dominant rDNA sequence in the genome can be used for phylogenetic reconstructions. Sequencing rDNA ITS regions following cloning, as was done in this analysis, sometimes leads to the detection of rare variants in the repeated rDNA sequences[35]. In order to determine whether the SNP pattern we have uncovered is a diagnostic species identifier or is due to the detection of rare cloning variants, we have begun an analysis using PCR-denaturing gradient gel electrophoresis in which rare rDNA variants from a single sample can be isolated and sequenced[36].
5. Conclusions
- Our results indicate the possibility that two reproductively isolated cryptic species that are independent of growth form, depth and reef location may exist off the coast of San Salvador, Bahamas. These results have important implications for using the paleontological record for investigating taxonomic and evolutionary relationships between closely related hydrozoan taxa. Inasmuch as macro- and microskeletal features are the only recourse for elucidating such relationships for fossil hydrozoans, we submit that a close examination of the facies in which specimens are preserved accompany their identification. In this fashion, patterns of ecophenotypic plasticity may be explored in tandem with taxonomic analysis. Many new Caribbean coral species and species complexes have been recognized by integrating molecular genetic and morphometric analyses[37-38] and these have been extended to include fossil corals[39]. However, the morphometric component of this work is carried out using landmark analyses of scleractinian coral skeletal microstructure. Hydrozoan skeletons are much less complex than those featured by scleractinians (for example no septa, much less septal ornamentation). It is possible that microstructural analyses of hydrozoans simply cannot yield results that mirror molecular genetic analyses.
ACKNOWLEDGEMENTS
- This work was conducted in the Bahamas under a permit granted by the Ministry of Agriculture and Marine Resources. We would like to thank Cornell College undergraduates Pavla Brachova, Peter Lehr, Arno Reichel and Halley Elledge for their assistance with the molecular data and numerous Cornell research classes for the abundance survey data. We would also like to thank Dr. Donald T. Gerace, Chief Executive Officer, and Dr. Tom Rothfus, Executive Director of the Gerace Research Centre, San Salvador, Bahamas. Completion of this work was made possible by Cornell College Faculty Development Grants (BG and CT), a Ringer Endowed Fellowship (CT), McElroy Research Grants (CT), and the LI-COR Biosciences Genomics Education Matching Funds Program (CT).
References
| [1] | Stearn, C.W., and R. Riding. 1973. Forms of the Hydrozoan Millepora on a recent coral reef. Lethaia 6:187-200. |
| [2] | Stoddart, D.R. 1969. Ecology and morphology of recent coral reefs. Biol Rev 44:433-498. |
| [3] | Edmunds, P.J. 1999. The role of colony morphology and substratum inclination in the success of Millepora alcicornis on shallow coral reefs. Coral Reefs. 18:133-140. |
| [4] | Boschma, H. 1948. The species problem in Millepora. Zool Verh Leiden 1:3-115. |
| [5] | Lewis, J.B. 1989. The ecology of Millepora. Coral Reefs 8:99-107. |
| [6] | Lewis, J.B. 2006. Biology and ecology of the hydrocoral Millepora on coral reefs. Advances in Marine Biology 50:1-55. |
| [7] | Gause, G.F. 1947. Problems of evolution. Trans Conn Acad Sci 37:17-68. |
| [8] | Foster, A.B. 1979. Phenotypic plasticity in the reef corals Montastrea annularis (Ellis and Solander) and Siderastrea siderea (Ellis and Solander). J Exp Mar Biol Ecol 39:25-54. |
| [9] | Brown, B.E., L. Sya’rani, and M.D. Le Tissier. 1985. Skeletal form and growth in Acropora aspera (Dana) from the Pulau Seribu, Indonesia. J Exp Mar Biol Ecol 86:139-150. |
| [10] | Palumbi, S.R. 1984. Tactics of acclimation: morphological changes of sponges in an unpredictable environment. Science 225:1478-1480. |
| [11] | Meyer, A. 1987. Phenotypic plasticity and heterochrony in Cichlasoma managuense (Pisces, Cichlidae) and their implications for speciation in cichlid fishes. Evol 41:1357-1369. |
| [12] | Lively, C.M. 1986. Predator-induced shell dimorphism in the acorn barnacle Chathamalus anisopoma. Evol 40:232-242. |
| [13] | Martín-Mora, E., F.C. James, and A.W. Stoner. 1995. Developmental plasticity in the shell of the queen conch Strombus gigas. Ecol 76:981-994. |
| [14] | Todd, P. 2008. Morphological plasticity in scleractinian corals. Biol Rev 83:315-337. |
| [15] | Weerdt, W.H.de 1981. Transplantation Experiments with Caribbean Millepora Species (Hydrozoa, Coelenterata), Including Some Ecological Observations on Growth Forms. Bijdragen Tot De Dierkunde 51:1-19. |
| [16] | Duchassaing De Fombressin, P., and J. Michelotti. 1864. Supplement Au Memoire Sur Les Coralliares Des Antilles. Mem R Acad Sci Torino 2:97-206. |
| [17] | Hickson, S.J. 1898. Notes on the collection of specimens of the genus Millepora obtained by Mr. Stanley Gardiner at Funafuti and Rotuma. Proc Zool Soc London. |
| [18] | Veron, J.E.N. 2000. In: Staffort-Smith, M. (Eds), “Corals of the World” vols 1-3. Australian Institute of Marine Science, Townsville, Australia. |
| [19] | Weerdt, W.H.de, and P.W. Glynn. 1991. A new and presumably now extinct species of Millepora (Hydrozoa) in the eastern Pacific. Zoologische Mededelingen 65:267-276. |
| [20] | Cairns, S.D., B.W. Hoeksema, and J. van der Land. 1999. Appendix: List of the extant stony corals. Atoll research Bulletin 459:13-46. |
| [21] | Hillis, D.M., and M.T. Dixon. 1991. Ribosomal DNA: molecular evolution and phylogenetic inference. Q. Rev. Biol. 66:411-453. |
| [22] | Schlotterer, C., M.T. Hauser, A. von Haeseler, and D. Tautz. 1994. Comparative evolutionary analysis of rDNA ITS regions in Drosophila. Molecular Biology and Evolution. 11:513-522. |
| [23] | Chen, C.A., B.L. Willis, and D.J. Miller. 1996. Systematic relationships between tropical corallimporharians (Cnidaria: Anthozoa: Corallimopharia): utility of the 5.8S and internal transcribed spacer (ITS) regions of the rRNA transcription unit. B Mar Sci 59(1):196-208. |
| [24] | Takabayashi, M., D. Carter, S. Ward, and O. Hoegh-Guldberg. 1998. Inter- and intra-specific variability in ribosomal DNA sequence in the internal transcribed spacer region of corals. Proc Aust Coral Reef Soc 75th Ann Conf 1:241-248. |
| [25] | Takabayashi, M., D.A. Carter, W.K.W. Loh, and O. Hoegh-Guldberg. 1998b. A coral specific primer for PCR amplification of the internal transcribed spacer region in ribosomal DNA. Mol Ecol 7:925-931. |
| [26] | Meroz-Fine, E., I. Brickner, Y. Loya, and M. Ilan. 2003. The hydrozoan coral Millepora dichotoma: Speciation or phenotypic plasticity? Mar Biol 143:1175-1183. |
| [27] | Online Available:http://earthobservatory.nasa.gov/IOTD/view.php?id=76134 |
| [28] | Amaral, F. D., M.K. Broadhurst, S.D. Cairns, and E. Schlenz. 2002. Skeletal morphometry of Millepora occurring in Brazil, including a previously undescribed species. Proceedings of the Biological Society of Washington 115 (3): 681-695. |
| [29] | Clarke, K. R., and R.M. Warwick. 1994. Change in marine communities: An approach to statistical analysis and interpretation. National Environment Research Council, UK. |
| [30] | Rowan, R., and D.A. Powers. 1991. Molecular genetic identification of symbiotic dinoflagelletes (Zooxanthellae). Mar Ecol Prog Ser 71:65-73. |
| [31] | Lopez, J.V., R. Kersanach, S.A. Rehner, and N. Knowlton. 1999. Molecular determination of species boundaries in corals: Genetic analysis of the Montastraea annularis complex using amplified fragment length polymorphisms and a microsatellite marker. Biology Bulletin, 196:80-93. |
| [32] | Odorico, D.M., and D.J. Miller. 1997. Internal and external relationships of the Cnidaria: implications of primary and predicted secondary structure of the 5’-end of the 23S-like rDNA. Proc R Soc Lond B Biol Sci 264:77–82. |
| [33] | Kumar S., M. Nei, J. Dudley, and K. Tamura 2008. MEGA: A biologist-centric software for evolutionary analysis of DNA and protein sequences. Briefings in Bioinformatics 9:299-306. |
| [34] | Vollmer, S.V. and S.R. Palumbi. 2004. Testing the utility of internally transcribed spacer sequences in coral phylogenetics. Mol. Ecol. 13:2763-2772. |
| [35] | LaJeunesse, T.C. and J.H. Pinzon. 2007. Screening intragenomic rDNA for dominant variants can provide a consistent retrieval of evolutionary persistent ITS (rDNA) sequences. Mol. Phylogenet. Evol. 45:417-422. |
| [36] | Muyzer, G., E. De Waal, and A.G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. |
| [37] | Budd, A. F., T.A. Stemann, and K.G. Johnson. 1994. Sratigraphic distributions of genera and species of neogene to recent Caribbean reef corals. J. Paleont. 68: 951-977. |
| [38] | Knowlton, N., J.C. Lang, and B.D. Keller. 1992. Sibling species in Montastrea annularis, coral bleaching and the coral climate record. Science 255: 330-333. |
| [39] | Budd, A. F., and K.G. Johnson. 1996. Recognizing species of Late Cenozoic Scleractinia and their evolutionary patterns, Paleontological Society Papers 1: 59-79. |
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-Text HTML
Full-Text HTML