-
Paper Information
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
Marine Science
p-ISSN: 2163-2421 e-ISSN: 2163-243X
2012; 2(2): 1-6
doi: 10.5923/j.ms.20120202.01
Isolation and Characterization of Gut Micro Biota from Some Estuarine Fishes
Kanagasabapathy Sivasubramanian , Samuthirapandian Ravichandran , R. Kavitha
Centre of Advanced Study in Marine Biology, Annamalai University, Parangipettai, 608502, India
Correspondence to: Kanagasabapathy Sivasubramanian , Centre of Advanced Study in Marine Biology, Annamalai University, Parangipettai, 608502, India.
| Email: |  |
Copyright © 2012 Scientific & Academic Publishing. All Rights Reserved.
Gut flora has a continuous and dynamic effect on the host's gut and systemic immune systems. The gut microbiota of fish has been studied extensively using various techniques; little information is available on the correlation between gut microbiota and host activity. In the present investigation three species of estuarine fishes were taken with regard to their microbial population in the gut region and enzymatic analysis of those gut isolates. The total heterotrophic bacterial load ranged from 0.8x106 to 4.5x106 CFU/g of gut sample and it was found to be the maximum of 4.5x106 CFU/g in Oreochromis mossambicus gut followed by 3.1x106 CFU/g in Oreochromis leucostictus gut and minimum of 0.8x106 CFU/g in Etroplus suratenis. The isolates were identified by various biochemical tests. Pseudomonas, Vibrio and Bacilli are the common bacteria found in all three fishes. The bacterial isolates were subjected to quantitative estimation of enzymes viz., amylase, lipase and protease. Varied kinds of results were obtained in different fish species. However it was observed that 20% of the isolates produced only amylase. A collection of 10 bacterial isolates from three fishes were screened for antagonistic activity against human and fish bacterial pathogens. Upon testing all these strains, it became apparent that strains isolated from the fish O.mossambicus, exhibited antimicrobial activity against Klebsiella pneumoniae (17mm) and Vibrio cholerae (12mm). Thus the present preliminary study showed the variation in the gut micro-flora of different fish species and the microbes from marine environment may be useful in development of probiotics, drugs and also in the industrial enzyme production.
Keywords: Estuarine fishes, Gut micro biota, THB, antimicrobial, Enzymes
Article Outline
1. Introduction
- Fish receive bacteria in the digestive tract from the aquatic environment through water and food that are populated with bacteria. Being rich in nutrient, the environment of the digestive tract of fish confers favorable conditions for the microorganisms. The importance of intestinal bacteria in the nutrition and well–being of their hosts has been established for homeothermic species, such as birds and mammals. However, there is limited information for fish, the poikil- othermic vertebrates. Gut micro flora plays an important role in the digestive process, growth and disease of the host. However some bacteria which possess the ability to tolerate the low pH in gastric juices resist the action of bile acids, lysozyme secreted in intestines, immune responses and adheres to the mucus or enteric wall surface could persist for a relatively long time and eventually make intestinal micro flora specific to each host animal[1]. It is clear that bacterial species presents in the gut can influence the health and robustness of the host. The influence of the gut flora on the host is clearly of great interest in aquaculture, particularly where poor productivity and/or stock losses are widespread. Within marine and other aquatic animals, the colonization of the digestive system by micro-organisms is influenced by a number of both host and non-host related factors. On the other hand, bacteria producing antibacterial substances were isolated from marine fish intestines[1,2] thereby suggesting that these bacteria may inhibit the establishment of invading bacteria in the fish intestine. However, it remains to be elucidated whether freshwater fish harbor the antibiotic-producers as well as marine fish. In the present study, an attempt has been made to investigate the variation of the gut bacterial species in the gastrointestinal (GI) tracts of three brackish water teleosts, namely the Oreochromis leucostictus, Oreochromis mossambicus, Etroplus suratenis. Further the intestinal isolates were evaluated for extracellular enzyme producing capacities and also examined for their antibacterial ability against fish and human pathogens.
2. Materials and Methods
2.1. Collection of Samples
- Fishes O. leucostictus, O. mossambicus, E. suratenis were collected from the Vellar estuary (Lat.11º29″ N and Long. 79º46″ E), South east coast of India and transformed to sterile polythene bags using sterile forceps. All the samples were transformed immediately to the laboratory and subjected to various analyses.
2.2. Isolation of the Gut and Their Homogenization
- Fishes were anaesthetized in an ice bath for 5–10 min and each individual surface sterilized by immersion for 30 seconds in 70% ethanol. The gut was aseptically dissected from the animal’s musculature. Gut was weighed and placed into a 10 ml sterile double strength phosphate-buffered saline (PBS) solution (disodium phosphate, 2-3% (w/v); sodium phosphate, 0-6% (w/v) and sodium chloride, 1-2% (w/v)). Gut was homogenized in tissue homogenizer.
2.3. Isolation and Identification of Bacteria
- One ml aliquot of the gut homogenate was aseptically spread with 9ml sterile double strength PBS onto Nutrient agar. All plates were incubated at 37℃ for 24-48hrs. Colonies developed on the plate were counted and expressed as cfu/g. The isolates were purified on the agar slants. The isolated bacterial strains were identified up to species level[3] and were stored in nutrient agar slants for further study. As an initial step in the identification of isolates, the following properties or activities were recorded: Gram stain, motility, oxidase activity, catalase activity, oxidation/ fermentation, glucose acid, glucose gas, pigment production and citrate utilization. A series of secondary tests was used, when required, to complete the genus level identification of isolates. These tests included growth on 0% sodium chloride media, production of amylase, production of protease, lipase activity and gelatinase activity.
2.4. Antagonistic Activity of Gut Isolates
- A spectrum of antibacterial activity was studied using test agent range of 10 different strains of human bacterial pathogen and 5 fish bacterial pathogenic gram positive and gram negative bacteria (Vibrio parahaemolyticus, Proteus mirabilis, Staphylococcus aureus, Vibrio cholera, Klebsiella pneumonia, Klebsiella oxytoca, Staphylococcus pyogens, Escherichia coli, Salmonella typhi, Salmonella paratyphi, Pseudomonas fluroscences, Aeromonas hydrophila, Vibrio alginolyticus, Enterobacter sp., Proteus sp). In vitro antibacterial assay was carried out by well diffusion technique[4]. The inhibition zone was measured and expressed in terms of diameter using caliper or a scale and recorded.
3. Results
3.1. Total heterotrophic Bacteria (THB)
- The numbers of cultivable bacterial cells present in fish gut were estimated after isolation and growth on nutrient agar (NA) plates incubated at room temperature at 37℃. The total heterotrophic bacterial load ranged from 1.8x106 to 4.8x106 CFU/g of gut sample and it was found to be the maximum of 4.5x106 CFU/g in O.mossambicus fish gut, 3.1x106 CFU/g in O.leucostictus fish gut and minimum of 0.8x106 CFU/g in E.suratenis (Fig 1).
 | Figure 1. THB load in various fish species |
|
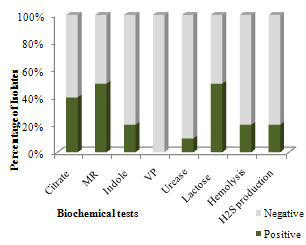 | Figure 2. Percentage of isolates showing biochemical activities |
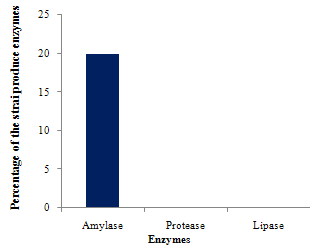 | Figure 3. Enzyme Production by the Isolates |
3.2. Antagonistic Activity of Gut Isolates
- The bacterial strains isolated from the gut of three fish species were assayed for their antimicrobial properties. Antimicrobial abilities were detected by well diffusion technique. A total of nine bacterial isolates were tested for their antagonistic ability against highly virulent 10 human bacterial and 5 fish bacterial pathogens. Maximum zone of inhibition was recorded against K.pneumoniae (17mm) and minimum against V.cholerae (12mm). No activity was recorded against the fish pathogens (Table 3).
|
4. Discussion
- In contrast to terrestrial animals, fishes have closer contact with the environmental micro-biota due to their aqueous habitat. Surrounding bacteria are continually ingested with food or water. For this reason, transient microorganisms probably have more constant and important interaction with fish gastrointestinal ecosystems compared to terrestrial animals. In general, genus Acinetobacter, Vibrio and Pseudomonas were the most common bacteria in the gut of marine fishes[5, 6, 7], and Vibrio was considered as one of the predominant bacteria[7, 8, 9, 10]. The experiment measured total bacterial numbers in the gastro intestinal region of three estuarine fishes. The percentage of isolated species and the percentage of isolate from any particular species varied significantly among gastro intestinal tract region. Aeromonas hydrophila, Aeromonas veronii were the most frequently isolated Gram negative bacilli[11]. In the present study, the micro-biota in the gut of three estuarine fishes was studied. The results showed that Acinetobacter, Vibrio and Pseudomonas were also isolated from the gut of fishes. Recently, Psychrobacter has been successfully isolated from the gut of marine fish, such as Arctic charr (Salvelinus alpinus)[12] and Atlantic cod (Gadus morhua)[13], but its role in the gut of fish is unclear. Pseudomonas sp. was isolated and dominated in the gut of all the three fishes used for present study. Therefore, the number and composition of gut micro-biota were discussed below. The total heterotrophic bacterial load ranged from 1.8x106 to 4.8x106 CFU/g of gut sample and it was found to be the maximum in O. mossambicus fish gut, and minimum in E.suratenis. Several factors, such as bacterial host specificity, food type and water resource[14] may explain these differences. Bacteria in the surrounding environment and feeding habit may have influence on the composition of the gastrointestinal microbiota in fish.O.mossambicus typically feed on the detritus which are rich in organic matter compared to the other fishes. O. leucostictus grazing on aquatic plants which are the rich in the starch content and E.suratenis naturally feeds on the tiny green plants (algae) present in the mid water rather than the bottom mud. It does not freely feed on large plant leaves. The feeding habitat is one of the factors may responsible for the presence of gut biota in these fishes. Both marine and freshwater fish have been shown to have a specific indigenous gut micro-biota[8, 15] and it may change with fish age, nutritional status, and environmental conditions[8]. The most common genera such as Vibrio, Pseudomonas and Acinetobacter were also isolated from the gut of E.suratenis. Interestingly, besides Gram positive Lactobacillus, Bacillus and several Gram negative bacteria normally in the gut of fish were also isolated and identified, such as Pseudomonas, Enterobacter and Flavobacterium. This normal indigenous flora can act competitively, excluding pathogens, inhibiting colonization and, consequently, preventing infection.Many workers have isolated bacteria which produce antibiotics from marine sources, including seawater, sediment; seaweed and fish gut[2,16,1,17]. In present study the isolates of estuarine fishes shows a broad range of activity against the pathogens. Maximum zone of inhibition was recorded against K.pneumoniae and minimum against V.cholerae. These results suggest that intestinal bacteria with antibacterial abilities may inhibit the growth of invading bacteria in intestines of freshwater fish, as well as marine fish, to some extent. The antibacterial effect of bacteria is generally due to any of the following factors, either singly or in combination: production of antibiotics, bacteriocins, siderophores, lysozymes or proteases, and alteration of pH values by the organic acids produced.[18] reported that Pseudomonas fluorescences reduced diseases caused by A. salmonicida.[19] also observed a similar phenomenon that Vibrio alginolyticus, used as a probiotic strain, reduced diseases caused by A. salmonicida, V. anguillarum and V. ordalii. Bacillus has been successfully isolated from the gut of several marine fish and applied as probiotics[20,21]. In the present study, Bacillus were the most dominant bacteria in the gut of these fishes and demonstrated antagonistic effect on pathogenic Vibrio (Table 3). Previous studies have showed that many Bacillus strains isolated from marine fish could inhibit potential pathogens[20,22] and one strain (Bacillus no. 48) has been successfully used as probiotics to exclude pathogenic Vibrio from common snook (Centropomus undecimalis)[20]. This work strongly suggests that effective control of microflora in fish intestines is possible using antibiotic-producing bacteria. A similar approach may be possible in freshwater fish using intestinal bacteria with an inhibitory effect against pathogenic bacteria[23]. Further studies along these lines should be undertaken in the near future in order to find an effective way of preventing outbreaks of infectious diseases mediated by cultured fish. Thus the present findings showed the variation in the micro-flora of different fish species and the microbes from marine environment might contribute to the incorporation of these bacteria in commercial aquaculture as supplement in formulated fish feed or in form of bacteria biofilm to achieve colonization in the fish gut at a higher degree and may be useful in development of probiotics, drugs as well as in the industrial enzyme production. However, further research involving potent bacterial strains should be conducted for evaluating their efficacy under actual farm conditions.
ACKNOWLEDGEMENTS
- The authors are grateful to thank Prof. T. Balasubramanian, our Dean, Centre of Advanced Study in Marine Biology, for his encouragement and Ministry of Environment & Forest (MoEn&F), Government of India for rendering a great financial support for conducting this research work.
References
| [1] | Olsson, J. C., Westerdhal, A., Conway, P. L., and Kjelleberg, S., 1992, Intestinal colonization potential of turbot (Scophthalumus maximus) and dap( Limanda limanda)- associated bacteria with inhibitory effects against Vibrio anguillarum. App. Envir.l micro., 58, 551-556. |
| [2] | A.M. Onarheim and J. Raa, Characterization and possible biological significance of autochthonous flora in the intestinal mucosa of sea water fish. In: Lesel R(ed) Microbiology in poecilotherms. Elsevier, Amsterdam, 1990. |
| [3] | R.E. Buchanan and N.E. Gibbons, Bergey's manual of determinative bacteriology, 8th edn. Williams & Wilkins Co. Baltimore, MD 1974. |
| [4] | Geels, F. P., and Schippers, G., 1983, Selection of antagonistic fluorescent Pseudomonas spp. and their root colonization and persistence following treatment of seed potatoes. J.patho., 108, 193-206. |
| [5] | Cahill, M. M., 1990, Bacterial flora of Fishes: A review. Micro. Eco., 19, 21-41. |
| [6] | Gatesoupe, F.J., Infante, J.L.Z., Cahu, C., and Quazuguel, P., 1997, Early weaning of sea bass larvae, Dicentrarchus labrax: the effect on microbiota, with particular attention to iron supply and exo-enzymes. Aquac., 158, 117-127. |
| [7] | Munro, P.D., Barbour, A., and Birkbeck, T.H., 1994, Comparison of the gut bacterial flora of start-feeding larval turbot reared under different conditions. J. App.Bact., 77, 560–566. |
| [8] | Olafsen, J.A., 2001, Interactions between fish larvae and bacteria in marine aquaculture. Aquac., 200, 223-247. |
| [9] | Eddy, S.D., and Jones, S.H., 2002, Microbiology of summer flounder Paralichthys dentatus fingerling production at a marine fish Hatchery. Aquac., 211, 9-28. |
| [10] | Sugita, H., and Ito, Y., 2006, Identification of intestinal bacteria from Japanese flounder (Paralichthys olivaceus) and their ability to digest chitin. Lett. Appl. Micro., 43, 336-342. |
| [11] | Molinari, L.M., Scoaris, D.O., Pedroso, R.B., Bittencourt, N.L.R., Nakamura, C.V., Ueda-Nakamura, T., Filho, B.A.A., and Filho, B.P.D., 2003, Bacterial microflora in the gastrointestinal tract of Nile tilapia,Oreochochromis niloticus, cultured in a semi-intensive system. Acta Scie. Biol. Sci., 25, 267–271. |
| [12] | Ringo, E., Sperstad, S., Myklebust, R., Mayhew, T.M., and Olsen, R.E., 2006a, The effect of dietary insulin on aerobic bacteria associated with hindgut of Arctic charr (Salvelinus alpinus L.). Aqua. Res., 37, 891-897. |
| [13] | Ringo, E., Sperstad, S., Myklebust, R., Refstie, S., and Krogdahl, A., 2006b, Characterisation of the microbiota associated with intestine of Atlantic cod (Gadus morhua L.) The effect of fish meal, standard soybeanmeal and a bioprocessed soybean meal. Aquac, 261, 829-841. |
| [14] | Verner-Jeffreys, D.W., Shields, R.J., Bricknell, I.R., and Birkbeck, T.H., 2003, Changes in the gut associated microflora during the development of Atlantic halibut (Hippoglossus hippoglossus L.) larvae in three Britishhatcheries. Aquac., 219, 21-42. |
| [15] | Vine, N.G., Leukes, W.D., and Kaiser, H., 2006, Probiotics in marine larviculture. FEMS Micro. Rev., 30, 404-427. |
| [16] | Westerdahl, A., Olsson, J.C., Kjelleberg, S., and Conway, P., 1991, Isolation and characterization of turbot (Scophrhalmus maimuskassociated bacteria with inhibitory effects against Vibrio unguillurum. Appl. Envi. Micro., 57, 2223-2228. |
| [17] | Romero Ayulo, A.M., Machado, R.A., and Scussel, V.M., 1994, Enterotoxigenic Escherichia coli and Staphylococcus aureus in fish and seafood from the southern region of Brazil. Int. J. Food .Microbio., 24,171-178. |
| [18] | Smith, P., and Davey, S., 1993, Evidence for the competitive exclusion of Aeromonas salmonicida from fish with stress-inducible furunculosis by a fluorescent pseudomonad. J. fish. dis., 16, 521-524. |
| [19] | Austin, B., Stuckey, L.F., Robertson, P.A.W., Effendi, I., Griffith, D.R.W., 1995, A probiotic strain of Vibrio alginolyticus effective in reducing diseases caused by Aeromonas salmonicida, Vibrio anguillarum and Vibrio ordalii. J. fish. dis., 18, 93-96. |
| [20] | Kennedy, S.B., Tucker, J.W., Neidic, C.L., Vermeer, G.K., Cooper, V.R., Jarrell, J.L., and Sennett, D.G., 1998, Bacterial management strategies for stock enhancement of warm water marine fish: a case study with common snook (Centropomus undecimalis). Bull. Mar. Sci., 62, 573-588. |
| [21] | Velmurugan, S., and Rajagopal, S., 2009, Beneficial uses of probiotics in mass scale production of marine ornamental fish. Afr. J. Micro. Res., 3, 185-190. |
| [22] | Sugita, H., Hirose, Y., Matsuo, N., and Deguchi, Y., 1998, Production of the antibacterial substance by Bacillus sp. strain NM 12, an intestinal bacterium of Japanese coastal fish. Aquac., 165, 269-280. |
| [23] | Sugita, H., Shibuya, K., Shimooka, H., and Deguchi, Y., 1996, Antibacterial abilities of intestinal bacteria in freshwater cultured fish. Aquac., 145, 195-203. |
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-Text HTML
Full-Text HTML
