-
Paper Information
- Next Paper
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
Journal of Microbiology Research
p-ISSN: 2166-5885 e-ISSN: 2166-5931
2025; 15(1): 1-6
doi:10.5923/j.microbiology.20251501.01
Received: Sep. 11, 2025; Accepted: Oct. 9, 2025; Published: Oct. 25, 2025

Identification of Microorganicms Isolated from Saline Soils and Their Agrobiological Significance
Ogiljon Jamalova Umarjonovna
Department of Biotechnology and Microbiology, Faculty of Biology and Ecology, M.Ulugbek National University of Uzbekistan, Tashkent, Uzbekistan
Correspondence to: Ogiljon Jamalova Umarjonovna, Department of Biotechnology and Microbiology, Faculty of Biology and Ecology, M.Ulugbek National University of Uzbekistan, Tashkent, Uzbekistan.
| Email: |  |
Copyright © 2025 The Author(s). Published by Scientific & Academic Publishing.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/

Background/Aim: The Aral Sea region faces extreme soil salinity due to environmental degradation, necessitating sustainable biological approaches. This study aimed to isolate salt-tolerant microorganisms, identify them, and evaluate their antifungal activity and agrobiological potential. Materials and Methods: Soil samples were collected in October 2024 from three depths (0–30 cm, 30–60 cm, 60–90 cm) in Uchsoy, Muynak district, Karakalpakstan. Microorganisms were isolated using serial dilution and cultured on various media (NA, Ashby, Chapek, Pikovskaya, Gauze, starch agar) Figure 1. Identification was performed via MALDI-TOF MS (Zybio EXS2600). Antifungal activity was tested using dual culture against Fusarium oxysporum, F. solani, and Alternaria spp. Soil pH, salinity, organic matter, and ion content were also analyzed. Results: Twenty-five strains were isolated; twelve were identified as Pseudomonas chlororaphis, Bacillus spp., Brevundimonas aurantiaca, Stenotrophomonas, Mucosa, and Aspergillus. Several strains tolerated up to 10% NaCl and showed strong antifungal effects, especially Pseudomonas and Bacillus species. Salinity and alkalinity influenced microbial diversity and activity. Conclusion: Native salt-resistant microorganisms possess significant antifungal potential and stress tolerance, making them suitable as biofertilizers and biocontrol agents for restoring saline-affected ecosystems in the Aral Sea region.
Keywords: Saline soil, Microorganicms, Maldi-tof, Agrobiological
Cite this paper: Ogiljon Jamalova Umarjonovna, Identification of Microorganicms Isolated from Saline Soils and Their Agrobiological Significance, Journal of Microbiology Research, Vol. 15 No. 1, 2025, pp. 1-6. doi: 10.5923/j.microbiology.20251501.01.
Article Outline
1. Introduction
- Currently, active research focuses on preserving soil fertility in highly saline, marshy, and sandy desert soils and on increasing biological productivity. Leading scientific centers and universities worldwide to work on developing short-rotation crop rotation systems, introducing biological preparations, cultivating various crops, and improving soil fertility. For example, a study by Zhang et al demonstrated that crop rotation enhances microbial diversity and promotes soil ecosystem multifunctionality. Several studies are also being conducted in our country. Specifically, efforts are focused on improving soil fertility in the dried Aral Sea basin, enhancing the ecological balance of agroecosystems, expanding the area of crops adapted to the soil and climate conditions of the desiccated Aral seabed, increasing land use efficiency, and reducing salt content in soils [1]. All of these tasks require the implementation of numerous practical measures aimed at mitigating the consequences of the Aral disaster, preserving the natural environment—including flora and fauna—for future generations, restoring vegetation in the Aral Sea basin, and creating protective forest plantations within the “Green Belt” initiative. "In the area around the Aral Sea, environmental problems, particularly the increase in soil salinity, have significantly decreased agricultural potential. Thus, the use of microorganisms to ensure ecological sustainability should be explored" [2]. The goal of this study is to isolate microorganisms from saline soils, investigate their biological properties, and evaluate their practical significance for agroecosystems.
2. Materials and Methods
2.1. Characteristics of the Study Area
- This study was conducted in the territory of the Uchsoy settlement (43.9016°N, 58.6803°E), located in the Muynak district of the Republic of Karakalpakstan. This region is an ecosystem crisis zone within the Aral Sea region, a consequence of the long-term drying of the sea, which has resulted in dry, saline, and hydromorphic soils [3]. The area has a sharply continental climate with an annual temperature range of -25℃ to +45℃ and receives only 80–100 mm of precipitation per year, the majority of which falls during the spring and winter [4]. These conditions lead to intense evaporation and a critical shortage of water resources.The soils in the Uchsoy area are predominantly sandy loam and sandy, with high salinity (EC) and a distinctly alkaline reaction (pH > 8). The primary types of saline soils are chloride-sulfate and carbonate, with concentrations varying by depth.Soil samples were collected from five points using the horizontal trench method at depths of 0-30 cm, 30-60 cm, and 60-90 cm. Following the ISO 10381 standard, samples weighing 500 to 1000 grams were taken from each depth, placed in plastic containers, and transported to the laboratory under refrigeration (ISO 10381-1:2002). For each layer, we determined the pH, electrical conductivity (EC), humus content, and microbiological composition [5].Environmental monitoring in the region has shown that soil salinization significantly reduces biological activity. However, these soils still host microorganisms adapted to such harsh conditions, particularly salt-resistant bacteria and fungi. The isolation, identification, and further study of these organisms offer promising potential for developing environmentally friendly biological products. Such products could enhance plant resistance to salinity, improve nutrient uptake, and ultimately contribute to the biological recovery and sustainability of agroecosystems [6,7,10].
2.2. Isolation of Microorganicms from Soil
- For the isolation and identification of microorganisms, the soil was initially subjected to serial dilutions using sterile water and sterile physiological saline solution (0.9% NaCl). To extract microorganisms from soil, including azotobacter, oligotrophs, actinomycetes and micromycetes, were grown on the particular solid selective nutrient media under conditions of isolation. Soil samples were collected for microbiological analysis. To accomplish this, 10 grams of soil were added to 90 milliliters of sterile water and then shaken for 10 minutes. Out of the suspension, 1 milliliter was taken with the help of a pipette. The remaining 9 milliliters of sterilized water was added to a test tube. The procedure of making a series of dilutions followed where the dilution factor was 1: 1 000 000 the whole process repeated three times to maintain the exactitude. Subsequently, in order to obtain isolates of single types of microbes of all generations, the right amount of suspension from each stage of dilution was inoculated onto the appropriate solid selective nutrient media through three replications, i.e., the process accounted for a total of twelve procedures of inoculation. The following culture media were used: meat peptone agar (MPA) for azotobacter, Ashby medium for oligotrophs, starch-ammonia agar (SAA) for actinomycetes, and Czapek-Dox medium for micromycetes. For all four kinds of media, the 4th dilution served as the inoculum, and the final determination was made after the growth of colonies. Cultures, seeded using the lawn plating method in Petri dishes, were incubated at a temperature of 28 ± 2°C for 2–7 days. Each colony was isolated based on morphological characteristics, purified, and then subjected to primary experiments using the Gram staining method [12,13,14].
2.3. Identification
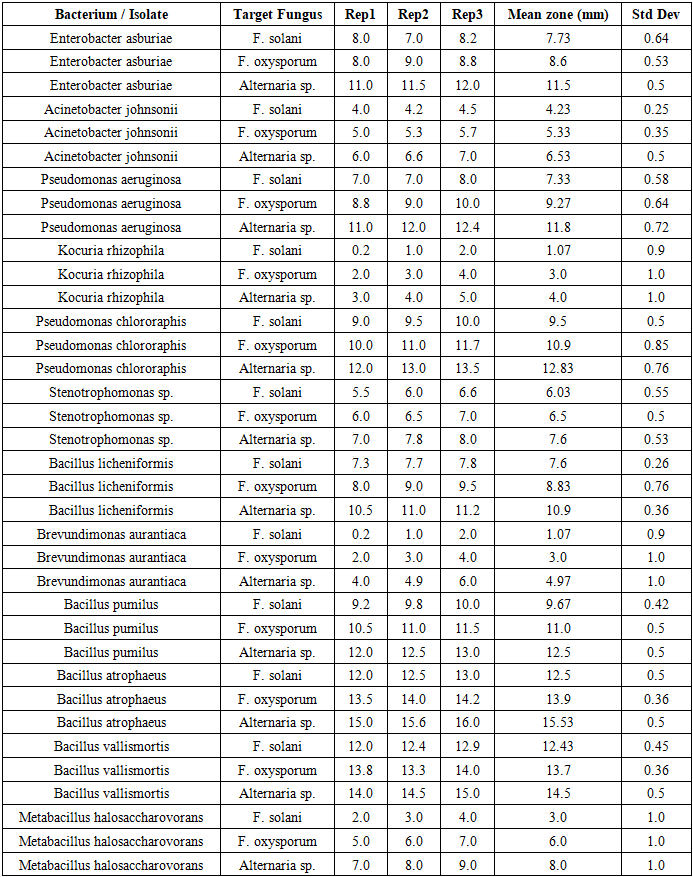
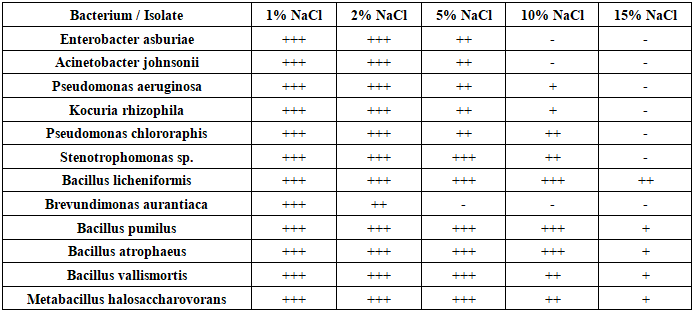
- The isolated microorganisms were identified using MALDI-TOF mass spectrometry (EXS2600, Zybio). Each pure colony was fixed in 70% ethanol, treated with formic acid and acetonitrile, and then subjected to spectral analysis by laser ionization. The identification results were processed using the MALDI Biotyper program, and based on the data obtained, the species, genus, and families of bacteria were identified [8]. The results of the study revealed the existence of several microorganisms, including Enterobacter asburiae, Acinetobacter johnsonii, Pseudomonas aeruginosa, rhizomatous cocuria (Kocuria rhizophila), Pseudomonas chlororaphis Pseudomonas aeruginosa, Stenotrophomonas sp., and lichen-like Bacillus licheniformis). The antagonistic activity of the isolated microorganisms against phytopathogenic fungi such as Fusarium oxysporum, Fusarium solani, and Alternaria spp. was studied by double cultivation (crop rotation) in Petri dishes. Each microorganicm and pathogen was simultaneously plated on the same nutrient medium. After 7 days, the growth zones were measured, and the antagonistic index (%) was calculated based on the size of the zones [9]. Determination of Soil Physicochemical Properties: The pH of the soil was determined using the potentiometric method, and the soil salinity was assessed by measuring electrical conductivity (EC). The humus content was determined using the Tyurin method (Tyurin). Average values were obtained from three replicates for each depth [15]. Antifungal Activity: The antagonistic activity of the isolated microorganisms against Fusarium oxysporum, Fusarium solani, and Alternaria was evaluated using the dual culture method in Petri dishes. Each colony was placed at the center of the dish, and the pathogenic fungus was seeded around it. The growth inhibition zones were measured by diameter (mm) Table 1. The antagonistic activity against Fusarium solani, Fusarium oxysporum, and Alternaria species, as well as the tolerance of microbial strains to NaCl at concentrations of 1%, 2%, 5%, 10%, and 15%, were studied and analyzed based on the data presented in Table 2.
|
|
3. Results
- During the study, 25 microorganisms were isolated, 12 of which were identified as follows: Some of the bacterial strains found include Enterobacter asburiae, infections caused by Acinetobacter johnsonii, Pseudomonas aeruginosa, Pseudomonas aeruginosa chlororaphisa, Stenotrophic bacillus, lichen bacillus, Brevundimona aurantiaca, Metabacillus halosaccharovorans, pumilus, atrophic bacillus, vallicmortis, Megatherium bacillus, a tiny stick. The main fungi include Mucor, Aspergillus niger, Aspergillus flavus and Penicillium chrysogenum Figure 2. Actinomycetes: Streptomyces roseus, Kocuria rhizophilus Figure 3.
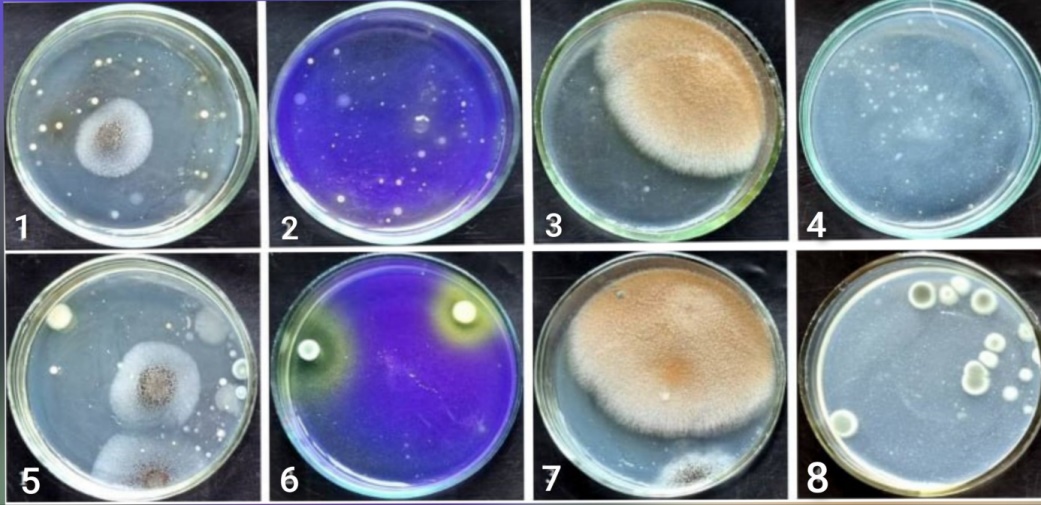 | Figure 1. Total count and morphology of the main physiological groups of microorganisms grown on different nutrient media at soil depths of 0 – 30 cm and 30 – 60 cm in the Aral Sea region |
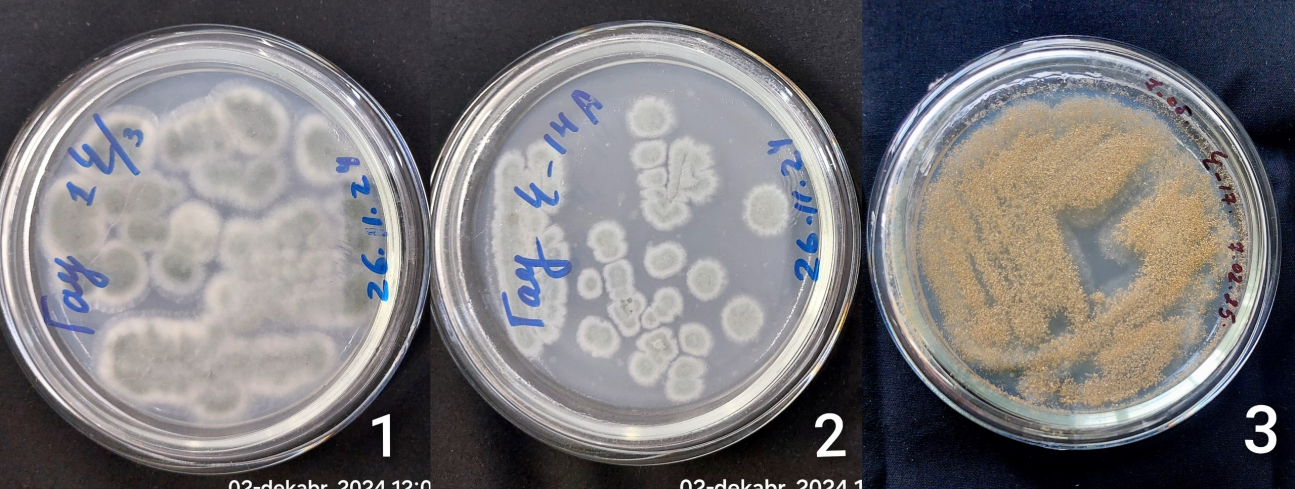 | Figure 2. Fungi isolated from saline soils in the research area |
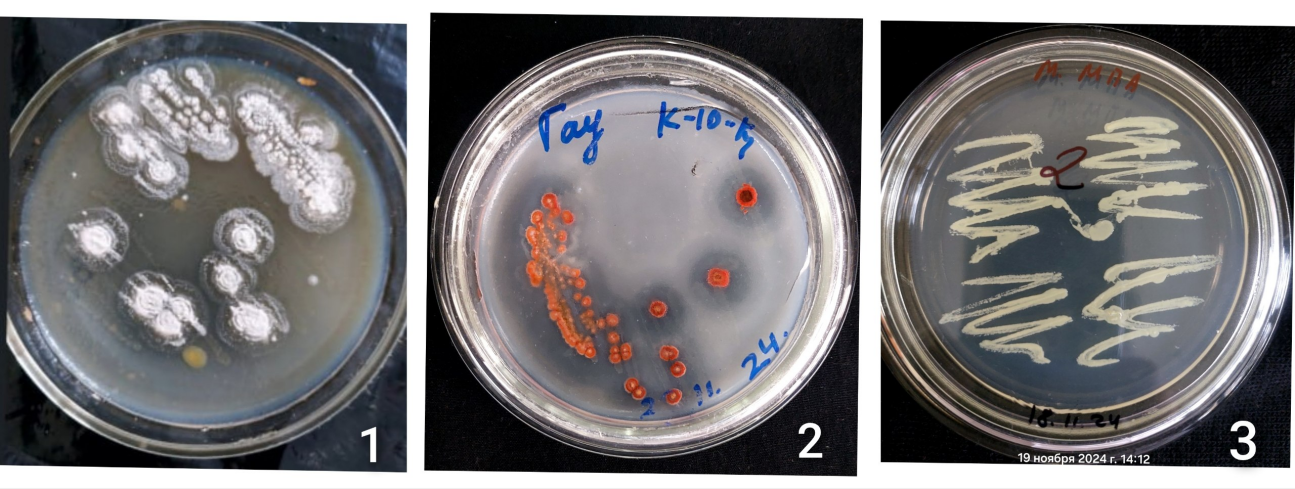 | Figure 3. Actinomycetes isolated from the research area |
4. Discussion
- The isolated microorganisms, among which the most active role was played by species of Bacillus and Pseudomonas chlororaphis, demonstrated high biological activity. Due to their antifungal and stress-resistant properties, they can become the main components in the production of bio-cork and bio-humus for saline soils [11]. The identification of salt-tolerant microbial strains forms the basis for the development of microbiological fertilizers adapted for these territories. Under conditions of strong salinization, such as in the Aral region, microorganisms developed their physiological and metabolic adaptation strategies. The ecological adaptation of salt-resistant microorganisms is of great agrobiological importance, as they contribute to improving plant growth, nutrition, and disease resistance. In particular, Bacillus and Pseudomonas species play an important role in the development of bio preparations due to their activity in the rhizosphere, production of siderophores, phosphate solubilization, and combat against phytopathogens [4].
5. Conclusions
- The results of this study showed that in saline soils of the Aral region, there are microorganisms with ecological and agrobiological significance. Based on soil samples taken at different depths in the Uchsoy area of the Moynak district, 12 species of microorganisms were isolated, which were identified using the MALDI-TOF mass spectrometry method. Among the isolated microorganisms, species such as Pseudomonas chlororaphis, Bacillus subtilis, Bacillus pumilus, Brevundimonas aurantiaca, Stenotrophomonas maltophilia, Aspergillus, and Mucor predominated. The isolated microorganisms are adapted to conditions of high salt content, and most of them showed high antagonistic reaction against phytopathogenic fungi. In particular, Pseudomonas and Bacillus species formed effective inhibition zones against Fusarium oxysporum, F. solani, and Alternaria spp. This demonstrates not only the biological role of these microorganisms as agents of biological control but also their potential as biofertilizers.The evaluation of the physicochemical properties of the soil at various depths, in combination with the microbial composition, confirms the direct influence of salinity and organic matter content on the diversity of microorganisms.
ACKNOWLEDGEMENTS
- The author(s) acknowledge the use of ChatGPT (OpenAI, GPT-4 version, 2025) during the preparation of this manuscript. Since English is not the author’s native language, the tool was used to assist with accurate translation into English and to check grammatical correctness and stylistic clarity. The content translated with the help of this tool was carefully reviewed and edited by the author(s), who take full responsibility for the final version of the publication.
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML