-
Paper Information
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
Journal of Microbiology Research
p-ISSN: 2166-5885 e-ISSN: 2166-5931
2022; 12(1): 13-23
doi:10.5923/j.microbiology.20221201.02
Received: Jan. 30, 2022; Accepted: Feb. 11, 2022; Published: Feb. 24, 2022

Incidence of Antibiotic Resistant Staphylococcus aureus in Leafy Greens and Clinical Sources
Cherinet Yigrem1, Broderick Eribo2, Kotu Sangre2
1Department of Biology, Howard University, 400 Bryan St NW Washington DC, USA
2Department of Biology, Howard University, 415 College St. NW Washington DC, USA
Correspondence to: Cherinet Yigrem, Department of Biology, Howard University, 400 Bryan St NW Washington DC, USA.
| Email: |  |
Copyright © 2022 The Author(s). Published by Scientific & Academic Publishing.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/

Staphylococcus aureus is a nosocomial pathogen associated with serious community and healthcare facility acquired infections. The current study determined the occurrence and diversity of antibiotic resistance Staphylococcus aureus in leafy vegetables sold in Gondar retail markets and clinical sources from Gondar community hospital. The microbiological analyses were performed based on standard methods. The vegetable samples were plated onto plate count agar and incubated for 48h at 30°C. The aerobic plate count of the vegetable samples ranged from 5.3 to 8.3 log10 cfu/g. Among the retail vegetable samples, 31 (28.18%) of them were positive for S. aureus and the average bacterial count was 8.85 cfu/g. The prevalence of S. aureus was highest in cauliflower (13/15, 86.7%), lettuce (13/15, 86.7%), followed by spinach (7/15, 46.7%), and cabbage (2/15, 13.4%). The isolates were analysed for antibiotics susceptibility test, and most isolates (99.3%) were resistant to vancomycin, ampicillin, penicillin, erythromycin, and methicillin. All the vegetable and clinical isolates (61, 100%) were multidrug resistant. There were no significant differences in phenotypic antibiograms between vegetable and clinical isolates (P=0.097>0.05). Phylogenetic tree determined the relationship between the vegetable isolates and the clinical isolates. Macrolide and beta lactam resistant genes and virulence factors were determined among the bacterial population. Four resistant genes were analysed in ten of the antibiotic resistant isolates using colony PCR and the results revealed that 90% were encoding (erm A), 100% (ermB), 100% (erm C), and 70% (msrA) resistant determinants. The present study also determined Hla, sea, Hlb, tst, and eta virulence factors and plasmids of the bacterial isolates.
Keywords: Antibiotic resistant, Resistance genes, Staphylococcus aureus, Plasmid, Virulence factors
Cite this paper: Cherinet Yigrem, Broderick Eribo, Kotu Sangre, Incidence of Antibiotic Resistant Staphylococcus aureus in Leafy Greens and Clinical Sources, Journal of Microbiology Research, Vol. 12 No. 1, 2022, pp. 13-23. doi: 10.5923/j.microbiology.20221201.02.
Article Outline
1. Introduction
- Staphylococcus aureus became one among the foremost common causes of nosocomial infections. Multiple antibiotic resistant Staphylococcus aureus pose a growing problem for human health globally [20,29]. The increase of antibiotic resistant virulent strains of Staphylococcus aureus, particularly methicillin resistant S. aureus (MRSA), may be a significant issue within the treatment and control of staphylococcal infections [61,37].Although, S. aureus is susceptible to known antibiotics [46,32,5], however, there are some antibiotic resistant strains [53,32]. Antibiotic resistance genes expressed by these strains are mainly acquired from external sources. This might be either natural or due to human actions mainly by antibiotic abuse, misuse and cause mutation and antibiotic selection [15,44]. The emergence and global dissemination of antibiotic resistant strains of S. aureus like methicillin resistant S. aureus (MRSA) is of health and socio-economic importance. Hence, antibiotic resistant strains are a major concern globally [26,44]. Annually, about 23, 000 people die due to antibiotic-resistant bacterial infections. Drug-resistant infections or “superbugs” could also cost a cumulative $100 trillion of economic output by 2050 if the world does not act to slow down the rise of drug resistance [7,38,34].One of the difficulties for therapeutic treatment of S. aureus infection is resistance to several commonly used antibiotics [7,38]. When S. aureus was first discovered, it had been easy to treat using available antibiotics. Meanwhile, after the discovery of penicillin sixty years ago to combat S. aureus, there have been strains of the pathogen that were immune to these antibiotics. A novel therapeutic agent called methicillin was developed and introduced to treat penicillin resistant S. aureus strains in 1961 [33]. The antibiotics, vancomycin, penicillin, and methicillin mode of action is extremely similar, and it involves inhibiting the synthesis of cell membrane through the stoppage of peptidoglycan formation by the pathogen and eventually lysis of the bacterium. However, a year after the introduction of methicillin, strains of S. aureus were reported to be methicillin resistant and gradually these strains disseminated worldwide. MRSA became a deadlier strain, which has become immune to most β- lactam antibiotics [19,56]. The antibiotic resistance of S. aureus is usually considered related to specific resistance genes. Resistance to macrolides (erythromycin and clindamycin) in staphylococci was supported by the active efflux mechanism encoded by the msrA gene, on the other hand resistant to β-lactams was supported by modification on 23S rRNA methylation encoded by ermA, ermB, and ermC genes [42,22]. Additionally, resistance of S. aureus to tetracycline was associated with tetM and tetK genes [48]. Staphylococcus aureus also, harbors genes encoding a variety of virulence factors including enterotoxins (SEs; sea to see, seg to sei, ser to set), exfoliative toxins (eta, etb), toxic shock syndrome toxin-1 (tst), Panton-Valentine leukocidin (lukS/F-PV), staphylococcal complement inhibitor (scn), and hemolysins (hly/hla, hlb, hld, hlgA, hlgB, hlgC) [72,73]. Enterotoxin production is important as it causes food poisoning, whereas the other virulence factors are associated with infection rather than intoxication. The scn gene is a marker of the immune evasion cluster (IEC) of strains that has been detected at high frequency among diverse collections of S. aureus strains obtained from humans, indicating that the scn gene may be useful for differentiating strains that have been transmitted from livestock to humans from those of human origin [74].Although there were handful of studies on antibiotic resistance S. aureus pathogens in Gondar Ethiopia, most of the articles focused on those pathogens isolated from meat, poultry, and milk in contrast, little information was found on vegetables [41]. Additionally, most of the works done regarding the incidence of antibiotic resistant bacteria are phenotypic, there are few molecular studies made in determining the resistant genes and virulence factors [21]. Therefore, the current study addressed the incidence and distribution of antibiotic resistant Staphylococcus aureus in leafy vegetables sold in Gondar retail markets related to clinical isolates from Gondar University Hospital. The article further determined whether leafy vegetables play a role as a carrier and reservoir of antibiotic resistant genes and identified virulence factors and plasmids among the bacteria population.
2. Materials and Methods
2.1. Sample Collection
- A total of 112 retail vegetable samples including cauliflower, spinach, lettuce, and cabbage, were collected from three different retail markets in Gondar, Ethiopia. The vegetable samples were placed in a cold box at a temperature approximately 4°C, tightly sealed with sterile plastic wrap, and transported to an accredited laboratory and subjected to microbiological analysis within 24 hrs. Among the total vegetable samples 31 of the isolates were confirmed as S. aureus. A total of 30 clinical strains were collected from Gondar university hospital. The isolates were identified from different specimens and no duplicate isolates from the same patient were included in this study. The organisms were identified to the species level using standard biochemical methods. S. aureus was identified by Gram staining, production of coagulase, catalase, DNase, and oxidation and fermentation of mannitol.The aerobic mesophilic count of S. aureus isolates in the vegetable samples were determined by the standard plate count method as described by the American Public Health association (APHA) on plate count agar [11]. Four tenfold serial decimal dilutions were made for each sample; 1 mL of each step was inoculated into duplicates of plate count agar (Himedia). For aerobic mesophilic plate count, the plates were incubated at 37°C for 24 h, and for the psychotropic plate count, incubated at 6°C for 5–7 days. The colonies formed on the plates were counted and expressed as log10 colony forming unit/g (log10 cfu/g) [43].
2.2. Antimicrobial Susceptibility
- To detect the S. aureus isolates for antibiotic susceptibility, the Kirby Bauer method [24] was performed by standard disk diffusion on Mueller–Hinton agar and incubated at 37°C for 24 h, following the guidelines of the Clinical and Laboratory Standards Institute [10]. A total of 25 antibiotics (Oxoid, Basingstoke, UK) were classified into 14 different groups [56]: amoxycillin/clavulanic acid (AMC, 30μg), ampicillin (AMP, 10 μg), cefepime (FEP, 10 μg), Cefoxitin (FOX, 30 μg), penicillin G (P, 10U), ceftazidime (CAZ, 30 μg), amikacin (AK, 30 μg), gentamicin (CN, 10 μg), kanamycin (K, 30 μg), streptomycin (S, 25 μg), chloramphenicol (C, 30μg), clindamycin (DA, 2μg), erythromycin (E, 15 μg), telithromycin (TEL, 15 μg), ciprofloxacin (CIP, 5 μg), Norfloxacin (NOR, 10μg), tetracycline (TE, 30μg), Linezolid (LZD, 30μg), rifampicin (RD, 5μg), Trimethoprim/sulphamethoxazole (SXT, 25 μg), Quinupristin/dalfopristin (QD, 15 μg), Teicoplanin(TEC, 30μg), Methicillin( 10μg), Nitrofurantoin (F, 300 μg) and Vancomycin (Van, 10 μg). Staphylococcus aureus ATCC 29213 and Escherichia coli ATCC25922 were included for quality control [11].
2.3. Plasmid Profile and 16s rRNA Sequencing of Bacteria Isolates
- The isolates were subjected to plasmid profile analysis via the OMEGA miniprep method. An overnight culture of every bacteria isolate was prepared in 5 mL of nutrient broth [44]. The broth culture was properly mixed by vertexing, and 1.5 mL was then transferred into a prelabeled Eppendorf tube. The tubes were then centrifuged for two minutes at 13 000 revolutions per minute (rpm) to reap the bacterial cells. The supernatant was gently decanted, leaving about 100 μL of the broth culture, which was then vortexed at high speed until the bacterial cell pellet became completely suspended [45]. OMEGA solution (300 μL; 25mM Tris, 10mM EDTA, 0.1N NaOH, 0.5% SDS) was then added to lyse the bacterial cells. The answer was mixed by inversion 3 for five times until the answer became slimy, after which 150 μL of three .0M sodium acetate (PH 5.2) was added and again vortexed for about 10 seconds [10]. The answer was centrifuged at 13 000 rpm for five minutes to discard cellular debris and chromosomal DNA [44]. The supernatant was then transferred into another labeled 1.5-mL Eppendorf tube, and 900 μL of cold absolute ethanol was added. The answer was centrifuged at 13 000 rpm for 10 minutes. The supernatant was discarded, and therefore the white pellet containing the plasmid DNA was rinsed twice with 1000 μL of 70% ethanol [26].The pellet was then air-dried, and 40 μL of TE buffer (10mM Tris, 1mM Na2EDTA) was added to the resuspend the pellet [45]. Electrophoresis of the extracted plasmid DNA was administered on 1.0% agarose gel and visualized with ethidium bromide via the ultraviolet transilluminator [45]. Selected and representative isolates were sent to GENWIZ for sequencing of the 16S rRNA genes.
2.4. Polymerase Chain Reaction (PCR) Amplification
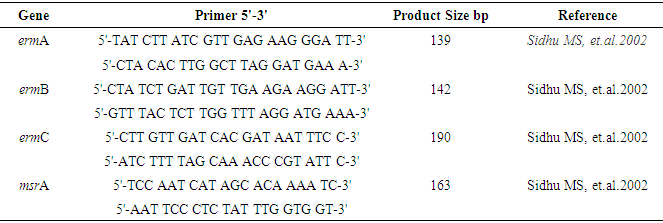
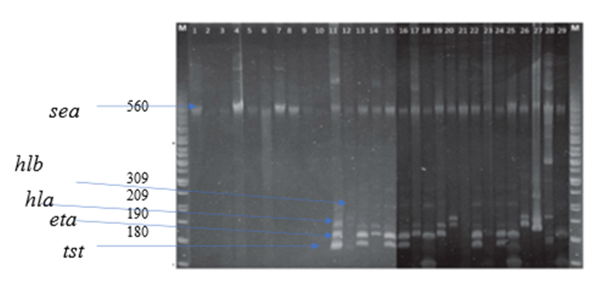
- Colony PCR was performed to detect all selected genes using the protocols listed in Table 1. The PCR mixture was obtained from EconoTaq PLUS and EconoTaq PLUS GREEN 2X Master Mixes, and each reaction was conducted under proper conditions: initial denaturation at 94°C for 4 minutes, followed by 35 cycles of 94°C for 45 seconds, polymerization at 72°C for 1 minute, and final extension for five minutes and 1 minute of annealing. Amplified products of all virulence factors are going to be further analyzed using ethidium bromide-mixed agarose gel (1.8%) under a UV Tran illuminator. Resistant genes were determined from the isolate supporting protocols listed in Table 1 and using standard primers (table 2) by performing colony PCR. Additionally, PCR was used to detect the presence of virulence genes: hla, hlb, sea, seb, sec, pvl, eta, etb, and tsst [34,22] mecA gene amplification was performed and identify MRSA isolates. SCCmec typing of MRSA strains was analyzed by multiplex PCR as previously described based on the primer’s conditions listed in table 3 [18].
|
2.5. Primers and PCR
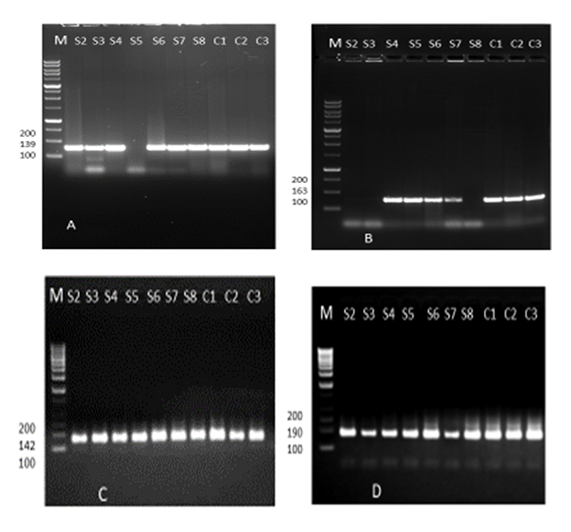
- The four primer pairs used to amplify the ermA, ermB, ermC, and msrA genes are listed in table 2 [51]. Within the current study, colony polymerase chain reaction (PCR) amplification was performed for ermA, ermC, and msrA genes as previously described [62] during a 25 µL reaction mixture consisting of: 1X PCR reaction buffer, 2.5 mM MgCl2, 0.2 mM dNTP, 25 per mol of every of the three primer pairs, 1.2 U of Taq DNA polymerase (Fermentas, Lithuania) and 200 ng DNA template. For ermB gene amplification, the ermB primers (25 per mol) were added during a separate PCR reaction under an equivalent above-mentioned condition. Amplification was administered employing a gradient thermal cycler (Bio-Rad, my cycler, US) because the following program: five minutes at 94°C, 30 cycles of amplification, consisting one minute at 95°C, 0.5 minutes at 55°C and two minutes at 72°C, ending with 10 minutes at 72°C (final extension). To verify amplification, 5 μL of PCR products were subjected to agarose gel electrophoresis (1.5% agarose, 1X TAE, 100V). Ethidium bromide-stained DNA fragments were then visualized with a UV transilluminator at 300 nm as compared to a 50bp/100bp molecular size standard ladder [21,16,13]. Additionally, PCR was used to detect the presence of nine virulence genes: hla, hlb, sea, seb, sec, pvl, eta, etb, and tsst [2,27,53]. All primers and the PCR amplification condition of resistance genes and virulence factors were listed in tables 2 and 3.
|
 | Table 3. Primers used for PCR of virulence factor detection |
2.6. Statistical Analysis
- To determine the aerobic mesophilic count among the vegetable samples and vegetable markets, serial dilution and log10 cfu/gm of colony counting time dilution factor divided by total volume were used. Differences in overall vegetable bacterial distribution among the vegetable types cauliflower, lettuce, spinach and cabbage were analyzed using average and percentage. Positive correlations in the antibiotic sensitivity tests between the bacterial isolates of the vegetable samples and clinical sources were assessed using one-way ANOVA. The z- tests were used to analyze the difference in resistance genes among the bacterial isolates. Significant differences in bacterial populations were observed across vegetable types using the nonparametric Kruskal-Willi’s test and between environmental samples and clinical sources employing a t-test.
3. Result
3.1. Aerobic Mesophilic Count
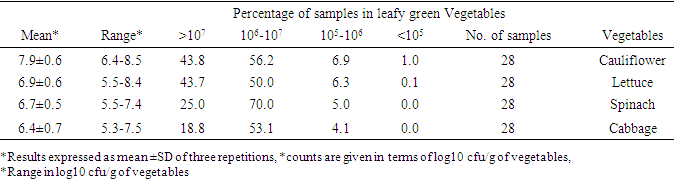
- The vegetable types, number of samples, counts of standard deviation (SD), and average (mean) in cfu/g of aerobic colony count are provided in table 4 below. The total bacterial count result indicated that all vegetable samples contain high bacteria load. The mean aerobic mesophilic count of the leafy vegetables was (7.9 cfu/g) cauliflower, (6.9 cfu/g) lettuce, (6.7 cfu/g) spinach and (6.5cfu/g) cabbage.
|
3.2. Antibiotic Susceptibility
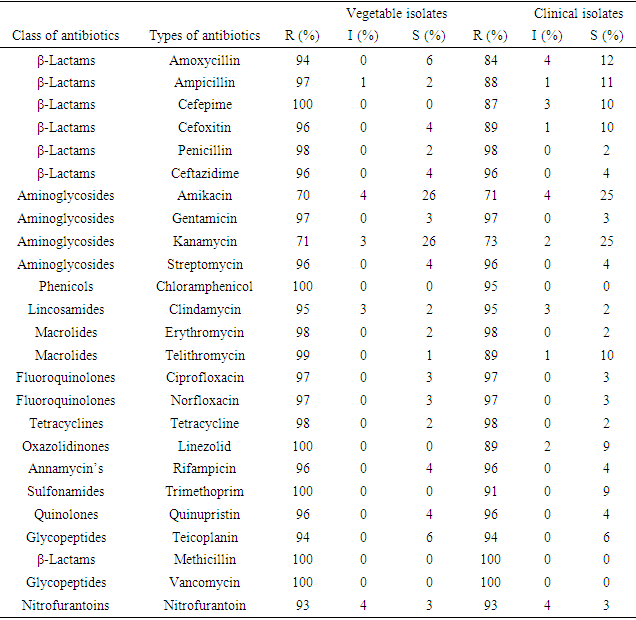
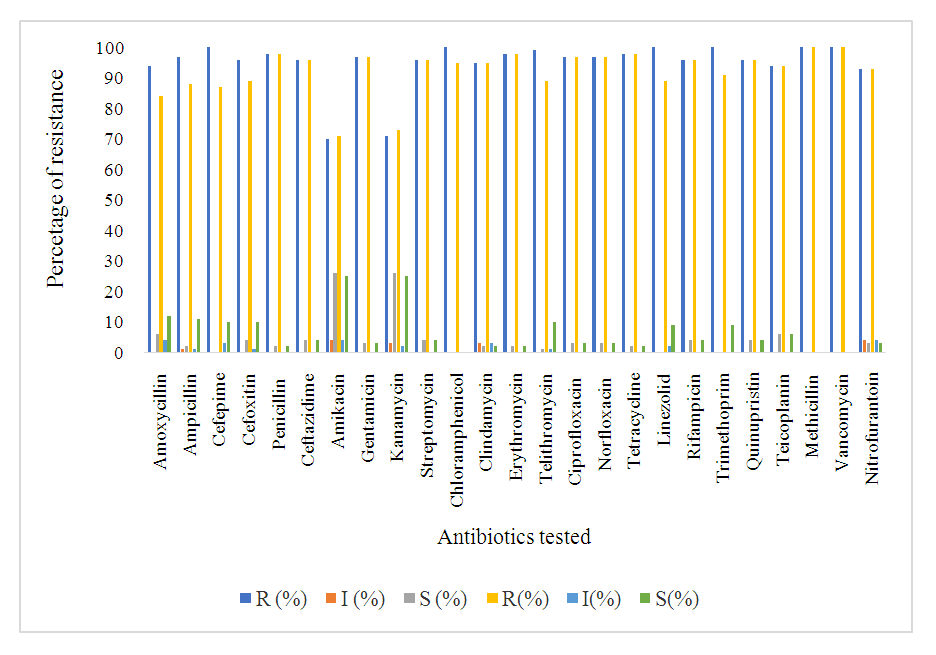
- The antimicrobial susceptibility patterns of Staphylococci isolated to various antibiotics are presented in table 3. Out of the total 31 vegetable isolates, 30(95.2%) exhibited resistance to penicillin, 31(100%) resistance to methicillin, and 31(100%) resistance to erythromycin. Most of these isolates were also resistant to tetracycline, levofloxacin, vancomycin, and gentamicin. Among the 30 clinical isolates of Staphylococci spp. 29(97%) were resistance to penicillin, 30(100%) resistance to erythromycin, and 30(100%) resistance to methicillin. Overall, each of the isolates showed multiple antibiotic resistant (resistant to more than three classes of antibiotics) in both vegetable and clinical isolates. Comparing the antibiogram of both sample sources shows similarity, P=0.097>0.05 (table 5 and figure 1).
|
3.3. Analysis of 16S rRNA gene Sequencing
- The sequences obtained in this study showed similarities between the sequences that obtained from clinical sources and sequences obtained from the vegetable samples respectively (MN872295, MN871695, MN871721, MN871820, MN872300, MN872311) clinical sources (MN871892, MN871924, MN871978, MN871721, and MN871988) vegetable samples as indicated in Fig. 2.The tree was constructed using the nucleotide sequences of eight 16s rRNA genes isolated from staphylococci isolates from clinical source and vegetable samples and one sequence obtained from the Gen bank as mentioned before (NR-118997.2). All sequences were clipped to an identical length corresponding to the same region before creating the tree (Fig. 2). The tree showed a high degree of similarity between all sequences of vegetable and clinical isolates or those imported from the Gen bank. The sequence of isolates (MN872295, MN871695, MN871721, MN871820, MN872300, and MN872311) that obtained from clinical sources, and sequences (MN871892, MN871924, MN871978, MN871721, and MN871988) obtained from vegetable samples respectively, are closer to each other and formed closed lineage (Figure 2).
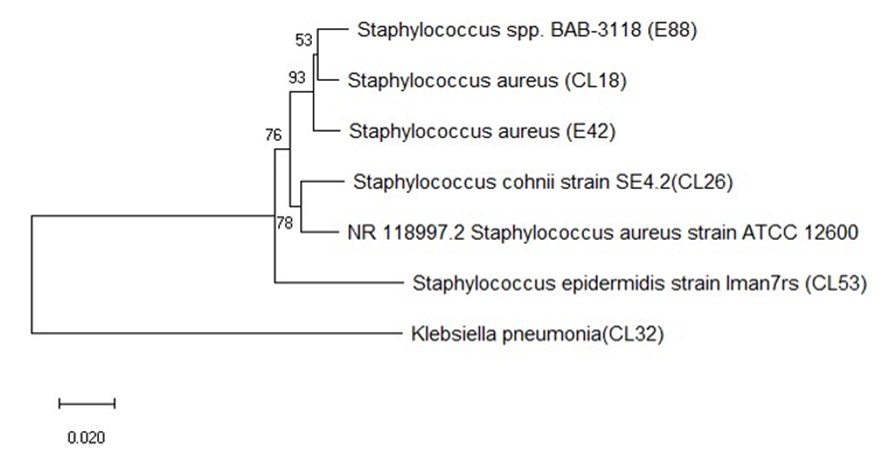 | Figure 2. Evolutionary relationship of 16s rRNA sequenced Staphylococci isolates (NR-118997.2 S. aureus strain ATCC12600 from NCBI) |
3.4. Plasmid Profiling of S. aureus Isolates
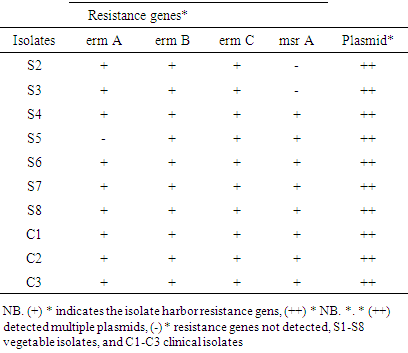
- Plasmid was isolated from both leafy vegetable isolates and clinical sources. Out of the twenty-one vegetable isolates, plasmids were detected in 13(61.9%) of the S. aureus isolates (Figure. 3A). The clinical isolates 10(100%) were found to have at least one plasmid (Figure 3. A1 & A2).
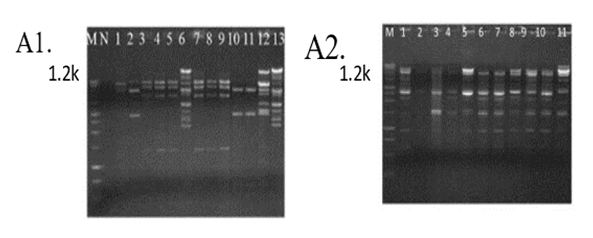 | Figure 3. Plasmid profile of Staphylococcus aureus A1) Vegetable and A2) Clinical isolates |
3.5. Analysis of Virulence Genes in Staphylococcus aureus Isolates
- Among the total (30) virulence factors screened isolates, the percentage of sea was highest (93.75) %), followed by hla (87.5) %), hlb (75.0) %), tst (62.5%), and eta (53.36%). Among the identified virulence genes, 90% of them exhibited no difference in frequency between MRSA and macrolide isolates of clinical origin (P > .05), except for the eta gene, which had a higher percentage in macrolides than in MRSA isolates (P < .05). The sea percentage was higher in MRSA strains and in macrolide strains among food isolates (P < .05). The Hla, Hlb, eta, and tst carriage rates among isolates of clinical origin were significantly higher than those among isolates of vegetable origin (P < .05). In contrast, the sea carriage rates were higher among both vegetable and clinical isolates (P < .05), Figure 4.
3.6. Analysis of the Macrolide and MRSA Resistance Gene Regions
- In this study, four different multiplex PCR reaction mixtures were defined for four resistance genes to identify macrolide (erythromycin) and β-lactam (MRSA) resistant genes among the bacteria population. The antibiogram results showed 30 (96.8%), 29(93.5%), and 31 (100%) isolates were resistant to penicillin, macrolide (erythromycin) and methicillin, respectively. A total of four available resistant genes were analyzed in 10 antibiotic-resistant isolates using colony polymerase chain reaction (CPCR). There were 9(90%) positive results of macrolide-resistant ermA, 10(100%) positive results of ermB, 10(100%) positive results of ermC, 7(70%) positive results of msrA were determined as depicted in Figures 5 (A), (B), (C), and (D) and table 6 below.
|
4. Discussions
- The primary focus of this study was to determine the total population and distribution of S. aureus in leafy vegetables and clinical sources from Gondar, Ethiopia including their antibiotic susceptibility, resistance genes, and other associated virulence factors. The results show that the vegetables had a high microbial load of Staphylococcus aureus [50].The aerobic mesophilic count of leafy greens, the mean aerobic mesophilic count of cauliflower, lettuce, spinach, and cabbage were 7.9 cfu/g, 6.9 cfu/g, 6.7 cfu/g, and 6.4 cfu/g, respectively, while the ranges were, cauliflower (6.4-8.5 cfu/g, lettuce (5.5-8.4 cfu/g) spinach (5.5-7.4 cfu/g) and cabbage (5.3-7.5 cfu/g). The higher bacterial load in this study may be partly due to the use of contaminated irrigation water and organic fertilizers in the farms, coupled with the poor hygienic environment where open defecation is not uncommon. Staphylococcus aureus is one among the foremost common pathogenic bacteria within the clinic and within the community, and therefore the presence or absence of resistance and virulence genes is closely associated with the pathogenicity of S. aureus infection. Investigation of the mechanisms of antibiotic resistance and detecting the distribution of resistance and virulence genes in S. aureus isolates from different sources will guide rational drug use among medical personnel’s and stop the dissemination of infection [39]. The antibiogram of the S. aureus isolates revealed that, 99% of vegetable and clinical S. aureus isolates were multidrug resistant. At present, our data is above previous reports among various food resources in other countries [47,60,8,9]. Majority of the isolates (98.9%) were resistant to erythromycin, vancomycin, methicillin, and penicillin, showing the research of leafy vegetables (96.3%) in South Korea [25]. Resistance to other antibiotics including tetracycline (98%,98%), clindamycin (95%,95%), gentamicin (97%,97%), and ciprofloxacin (97%,97%) clinical and vegetable respectively, was also higher than the previous report, 49.4% of S. aureus isolates from food products were resistant to tetracycline, 24.1% to clindamycin, and 13.8% to gentamicin [14]. Considering these antibiotics are increasingly utilized in animal breeding or human treatment and exchange of antibiotic-resistant genes by mobile genetic elements (MGEs) [36], it is not surprising that resistant strains become more common within the present. As we all know, methicillin resistant S. aureus (MRSA) represents a significant public health issue due to its ability to colonize and infect humans and animals among the isolates of S. aureus during this study the antibiogram showed that each one was MRSA strains. Cross contamination from environments could also be the main reason because animal-derived food products are widely known to be a crucial reservoir for MRSA [60]. However, the high antimicrobial resistance of S. aureus observed during this study should receive much attention. Macrolide and beta-lactam resistant gene regions (erm A, erm B, erm C, and misr A) were detected among the clinical and vegetable S. aureus isolates. Macrolide (erythromycin) resistance in staphylococci is encoded by erm gene [67]. All isolates found to be resistant to erythromycin by phenotypic methods contained a minimum of one erythromycin resistance gene. Our study confirmed that erm A, erm B, and msrA genes in S. aureus isolates were found at higher rates than in coagulase negative staphylococci, and among the coagulase negative staphylococci, ermC gene was more prevalent which agreed with previous reports [58]. In ten isolates, both erm(B) nor erm(C) gene was detected, although these were detected as resistant. Erythromycin resistance could be due to the presence of msrA or ermB genes as previously described in coagulase negative staphylococci [68]. Lim et al., reported that the ermA gene was more predominant than the opposite erythromycin resistance genes in S. aureus isolates, and the ermC gene was found mostly in coagulase negative staphylococci [69]. Similarly, during a study performed by Martineau et al., the ermC gene has been reported to be more prevalent in coagulase negative staphylococci. However, ermA has been reported to be the more common gene in coagulase negative in another study [70].The isolates were also screened for virulence genes to work out their pathogenic risk. All clinical strains were positive for Hla, in accordance with a study conducted in Asturias, which found that each one strain isolated from cow milk harbored the Hla virulence gene [23]. Moreover, it had been reported that Hla was associated with an epidemic of staphylococcal gastrointestinal disorders [35]. Another important factor underlying staphylococcal gastrointestinal disorder is that the presence of enterotoxins, although the share of enterotoxin genes in isolates of food origin varies in several studies. Different levels of virulence genes indicate different pathogenic potentials [35]. This current study showed that sea was the highest (93.75) %), followed by hla (87.5 %), hlb (75.0) %), tst (62.5%), and eta (53.36%); these percentages were above those identified in chicken products from Egypt, [17] indicating that the genes may significantly contribute to staphylococcal gastrointestinal disorder.The transmission of bacterial virulence factors to humans through the food chain as well as the higher multidrug resistance rates in those strains represents large risks for human infection. When resistance gene carriage rates were compared between MRSA and macrolide strains of food origin, only the etb rate was higher in MRSA strains; other genes demonstrated no differences. Combined with the results obtained for clinical isolates, our study suggested that the difference in virulence gene carriage rates between MRSA and macrolide strains might be insignificant [71].
5. Conclusions
- In this study we have analyzed the microbial load of four different leafy greens in Gondar town at the point of retail sale. Our study takes a combined bioinformatical approach to acquire detailed data on antimicrobial resistant and virulence markers in S. aureus isolated from retail vegetables and clinical sources from Gondar community hospital. MRSA and Macrolide resistant isolates were detected among all the isolates. Resistance genes towards β-lactam antimicrobials, tetracycline, and erythromycin were frequently observed, which correlates with different antimicrobial classes being commonly used in Gondar clinical cares. In addition, common virulence gene were detected among the isolates. Generally, our results show that both clinical and vegetable isolates are resistant to all antibiotics. Due to the presence of antibiotic resistant S. aureus, as well as other opportunistic and emerging pathogens in the examined fresh vegetables and clinical sources, this study emphasizes the need for a new guideline to address this public health nuisance. In addition, as antibiotic resistance becomes a clear public health problem in the area, there needs to be closer monitoring by the health department.
ACKNOWLEDGEMENTS
- I would like to express my sincere gratitude to Gondar University Department of Microbiology and Immunology and Howard University Biology department, applied microbiology laboratory.
Conflict of Interest
- There is no conflict of interest with any party in this research work.
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML