| [1] | A. Abdi-Ali, M. Mohammadi-Mehr, Y. Agha Alaei, Bactericidal activity of various antibiotics against biofilm-producing Pseudomonas aeruginosa, International Journal of Antimicrobial Agents, Volume 27, Issue 3, 2006, Pages 196-200. |
| [2] | Akeredolu DO., Akinnibosun FI., Udochukwu U. (2017). Isolation and identification of plasmids of bacteria from petroleum products contaminated soil. Journal of Bacteriology & Mycology, Volume 5 Issue 6 – 2017. |
| [3] | Akoachere, J., Tatsinkou, B. F., & Nkengfack, J. M. (2018). Bacterial and parasitic contaminants of salad vegetables sold in markets in Fako Division, Cameroon and evaluation of hygiene and handling practices of vendors. BMC research notes, 11(1), 100. https://doi.org/10.1186/s13104-018-3175-2. |
| [4] | Altschuler M, Heddens DK, Diveley RR Jr, Kresheck GC (1994) Plasmid DNA isolation utilizing a novel nonionic detergent. Biotechniques 17: 434–436. |
| [5] | Balasubramanian, D., Schneper, L., Kumari, H., & Mathee, K. (2013). A dynamic and intricate regulatory network determines Pseudomonas aeruginosa virulence. Nucleic acids research, 41(1), 1–20. https://doi.org/10.1093/nar/gks1039. |
| [6] | Bennett P. M. (2008). Plasmid encoded antibiotic resistance: acquisition and transfer of antibiotic resistance genes in bacteria. British journal of pharmacology, 153 Suppl 1(Suppl 1), S347–S357. https://doi.org/10.1038/sj.bjp.0707607. |
| [7] | Birnboim, H. C., & Doly, J. (1979). A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic acids research, 7(6), 1513–1523. https://doi.org/10.1093/nar/7.6.1513. |
| [8] | Bruno TF, Buser DE, Syme RM, Woods DE, Mody CH., (1998). Pseudomonas aeruginosa exoenzyme S is a mitogen but not a superantigen for human T lymphocytes. Infect Immun. 1998; 66:3072-3079. |
| [9] | C. Van Delden, B.H. Iglewski. Cell-to-cell signaling and Pseudomonas aeruginosa infections. Emerg Infect Dis, 4 (1998), pp. 551-560. |
| [10] | C.T. O'Loughlin, L.C. Miller, A. Siryaporn, K. Drescher, M.F. Semmelhack, B.L. Bassler, A quorum-sensing inhibitor blocks Pseudomonas aeruginosa virulence and biofilm formation Proc Natl Acad Sci U S A, 110 (2013), pp. 17981-17986. |
| [11] | Chowdhury EH, Akaike T (2005) Rapid isolation of high quality, multimeric plasmid DNA using zwitterionic detergent. J Biotechnol 119: 343–347. |
| [12] | Chowdhury K (1991) One step 'miniprep' method for the isolation of plasmid DNA. Nucleic Acids Res 19: 2792. |
| [13] | Currier TC, Nester EW (1976) Isolation of covalently closed circular DNA of high molecular weight from bacteria. Anal Biochem 76: 431–441. |
| [14] | D.E. Woods, S.J. Cryz, R.L. Friedman, B.H. Iglewski Contribution of toxin A and elastase to virulence of Pseudomonas aeruginosa in chronic lung infections of rats. Infect Immun, 36 (1982), pp. 1223-1228. |
| [15] | Donlan, R. M., & Costerton, J. W. (2002). Biofilms: survival mechanisms of clinically relevant microorganisms. Clinical microbiology reviews, 15(2), 167–193. https://doi.org/10.1128/CMR.15.2.167-193.2002. |
| [16] | E. Déziel, Y. Comeau, R. Villemur. Initiation of biofilm formation by Pseudomonas aeruginosa 57RP correlates with emergence of hyperpiliated and highly adherent phenotypic variants deficient in swimming, swarming, and twitching motilitiesJ Bacteriol, 183 (2001), pp. 1195-1204. |
| [17] | Fazeli N, Momtaz H., (2014). Virulence gene profiles of multidrug-resistant Pseudomonas aeruginosa isolated from Iranian hospital infections. Iran Red Crescent Med J. 2014; 16: e15722. |
| [18] | Finlayson EA, Brown PD., (2011). Comparison of antibiotic resistance and virulence factors in pigmented and non-pigmented Pseudomonas aeruginosa. West Indian Med J. 2011; 60:24-32. |
| [19] | Flores-Díaz, M., Monturiol-Gross, L., Naylor, C., Alape-Girón, A., & Flieger, A. (2016). Bacterial Sphingomyelinases and Phospholipases as Virulence Factors. Microbiology and molecular biology reviews: MMBR, 80(3), 597–628. https://doi.org/10.1128/MMBR.00082-15. |
| [20] | Fricks-Lima J, Hendrickson CM, Allgaier M, Zhuo H, Wiener-Kronish JP, Lynch SV, Yang K. (2011). Differences in biofilm formation and antimicrobial resistance of Pseudomonas aeruginosa isolated from airways of mechanically ventilated patients and cystic fibrosis patients. Int J Antimicrob Agents. 2011 Apr; 37(4): 309-15. |
| [21] | Ghanbarzadeh Corehtash Z, Khorshidi A, Firoozeh F, Akbari H, Mahmoudi Aznaveh A., (2015). Biofilm formation and virulence factors among Pseudomonas aeruginosa isolated from burn patients. Jundishapur J Microbiol; 8: e22345. |
| [22] | Guerry P, LeBlanc DJ, Falkow S (1973) General method for the isolation of plasmid deoxyribonucleic acid. J Bacteriol 116: 1064–1066. |
| [23] | Gultie, A., & Sahile, S. (2013). Microbial Spectrum of Fruit in Gondar town Markets, North Western Ethiopia. Journal of Microbiology Research, 3, 1-10. |
| [24] | Hassuna, N. A., Mandour, S. A., & Mohamed, E. S. (2020). Virulence Constitution of Multi-Drug-Resistant Pseudomonas aeruginosa in Upper Egypt. Infection and drug resistance, 13, 587–595. https://doi.org/10.2147/IDR.S233694. |
| [25] | Head NE, Yu H., (2004). Cross-sectional analysis of clinical and environmental isolates of Pseudomonas aeruginosa: biofilm formation, virulence, and genome diversity. Infect Immun; 72:133-144. |
| [26] | I. Mitov, T. Strateva, B. Markova. Prevalence of virulence genes among Bulgarian nosocomial and cystic fibrosis isolates of Pseudomonas aeruginosa, Braz J Microbiol, 41 (2010), pp. 588-595. |
| [27] | Iglewski BH. Pseudomonas. In: Baron S, editor. Medical Microbiology. 4th edition. Galveston (TX): University of Texas Medical Branch at Galveston; 1996. Chapter 27. Available from: https://www.ncbi.nlm.nih.gov/books/NBK8326/. |
| [28] | Ilori MON, Amund DL. Degradation of anthracene by bacteria isolated from oil polluted tropical soils. Z Naturforsch. 2000; 55(11–12): 890–897. |
| [29] | Itah AY, Essien JP., (2005). Growth profile and hydrocarbonoclastic potential of microorganisms isolated from tarballs in the bight of bonny. Nigeria. World J Microbial Biotechnol. 2005; 21(6–7): 1317–1322. |
| [30] | Jaffe, R. I., Lane, J. D., & Bates, C. W. (2001). Real-time identification of Pseudomonas aeruginosa direct from clinical samples using a rapid extraction method and polymerase chain reaction (PCR). Journal of clinical laboratory analysis, 15(3), 131–137. https://doi.org/10.1002/jcla.1016. |
| [31] | Jimenez, P. N., Koch, G., Thompson, J. A., Xavier, K. B., Cool, R. H., & Quax, W. J. (2012). The multiple signaling systems regulating virulence in Pseudomonas aeruginosa. Microbiology and molecular biology reviews: MMBR, 76(1), 46–65. https://doi.org/10.1128/MMBR.05007-11. |
| [32] | John, J. F., Jr, & Twitty, J. A. (1986). Plasmids as epidemiologic markers in nosocomial gram-negative bacilli: experience at a university and review of the literature. Reviews of infectious diseases, 8(5), 693–704. https://doi.org/10.1093/clinids/8.5.693. |
| [33] | Jurado-Martín I, Sainz-Mejías M, McClean S. Pseudomonas aeruginosa: An Audacious Pathogen with an Adaptable Arsenal of Virulence Factors. International Journal of Molecular Sciences. 2021; 22(6): 3128. https://doi.org/10.3390/ijms22063128. |
| [34] | Khalil MA, Ibrahim Sonbol F, Mohamed AF, Ali SS., (2015). Comparative study of virulence factors among ESbetaL-producing and nonproducing Pseudomonas aeruginosa clinical isolates. Turk J Med Sci; 45:60-69. |
| [35] | Khattab MA, Nour MS, El Sheshtawy NM. Genetic identification of Pseudomonas aeruginosa virulence genes among different isolates. J Microb Biochem Technol. 2015; 7:274-277. |
| [36] | L.R. Beuchat. Ecological factors influencing survival and growth of human pathogens on raw fruits and vegetables. Microbes and Infection, 4 (4) (2002), pp. 413-423 |
| [37] | LaSarre, B., & Federle, M. J. (2013). Exploiting quorum sensing to confuse bacterial pathogens. Microbiology and molecular biology reviews: MMBR, 77(1), 73–111. https://doi.org/10.1128/MMBR.00046-12 |
| [38] | Lezin, G., Kosaka, Y., Yost, H. J., Kuehn, M. R., & Brunelli, L. (2011). A one-step miniprep for the isolation of plasmid DNA and lambda phage particles. PloS one, 6(8), e23457. https://doi.org/10.1371/journal.pone.0023457. |
| [39] | Limoli, D. H., Jones, C. J., & Wozniak, D. J. (2015). Bacterial Extracellular Polysaccharides in Biofilm Formation and Function. Microbiology spectrum, 3(3), 10.1128/microbiolspec. MB-0011-2014. https://doi.org/10.1128/microbiolspec.MB-0011-2014. |
| [40] | Lister, P. D., Wolter, D. J., & Hanson, N. D. (2009). Antibacterial-resistant Pseudomonas aeruginosa: clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clinical microbiology reviews, 22(4), 582–610. https://doi.org/10.1128/CMR.00040-09. |
| [41] | M.A. Khattab, M.S. Nour, N.M. El Sheshtawy Genetic identification of Pseudomonas aeruginosa virulence genes among different isolates. J Microb Biochem Technol, 7 (2015), pp. 274-277. |
| [42] | Meletis G. (2016). Carbapenem resistance: overview of the problem and future perspectives. Therapeutic advances in infectious disease, 3(1), 15–21. https://doi.org/10.1177/2049936115621709. |
| [43] | Moazami Goudarzi S, Eftekhar F. Assessment of carbapenem susceptibility and multidrug resistance in Pseudomonas aeruginosa burn isolates in Tehran. Jundishapur J Microbiol. 2013; 6(2): 162–5. |
| [44] | Moges, F., Endris, M., Mulu, A., Tessema, B., Belyhun, Y., Shiferaw, Y., Huruy, K., Unakal, C., & Kassu, A. (2014). The growing challenges of antibacterial drug resistance in Ethiopia. Journal of global antimicrobial resistance, 2(3), 148–154. https://doi.org/10.1016/j.jgar.2014.02.004. |
| [45] | Moradali, M. F., Ghods, S., & Rehm, B. H. (2017). Pseudomonas aeruginosa Lifestyle: A Paradigm for Adaptation, Survival, and Persistence. Frontiers in cellular and infection microbiology, 7, 39. https://doi.org/10.3389/fcimb.2017.00039. |
| [46] | Nikbin VS, Abdi-Ali A, Feizabadi MM, Gharavi S. Pulsed field gel electrophoresis & plasmid profile of Pseudomonas aeruginosa at two hospitals in Tehran, Iran. Indian J Med Res. 2007; 126(2): 146–51. |
| [47] | Norouzi F, Mansouri S, Moradi M, Razavi M., (2010). Comparison of cell surface hydrophobicity and biofilm formation among ESBL-and non-ESBL-producing Pseudomonas aeruginosa clinical isolates. Afr J Microbiol Res; 4:1143-1147.of Foods and Beverages, Food Microbiology and Food Safety, DOI 10.1007/978-1-4419-0826-1-6. |
| [48] | P. Lanotte, S. Watt, L. Mereghetti, N. Dartiguelongue, A. Rastegar-Lari, A. Goudeau, et al. Genetic features of Pseudomonas aeruginosa isolates from cystic fibrosis patients compared with those of isolates from other origins. J Med Microbiol, 53 (2004), pp. 73-81. |
| [49] | P.R. Jacome, L.R. Alves, A.B. Cabral, A.C. Lopes, M.A., (2012). Maciel. Phenotypic and molecular characterization of antimicrobial resistance and virulence factors in Pseudomonas aeruginosa clinical isolates from Recife, State of Pernambuco, Brazil. Rev Soc Bras Med Trop, 45 (2012), pp. 707-712. |
| [50] | Pachori, P., Gothalwal, R., & Gandhi, P. (2019). Emergence of antibiotic resistance Pseudomonas aeruginosa in intensive care unit; a critical review. Genes & diseases, 6(2), 109–119. https://doi.org/10.1016/j.gendis.2019.04.001. |
| [51] | Paul B, Cloninger C, Felton M, Khachatoorian R, Metzenberg S (2008) A nonalkaline method for isolating sequencing-ready plasmids. Anal Biochem 377: 218–222. |
| [52] | Pearson, J. P., Feldman, M., Iglewski, B. H., & Prince, A. (2000). Pseudomonas aeruginosa cell-to-cell signaling is required for virulence in a model of acute pulmonary infection. Infection and immunity, 68(7), 4331–4334. https://doi.org/10.1128/IAI.68.7.4331-4334.2000. |
| [53] | Prasad, A.S.B., Shruptha, P., Prabhu, V. et al. Pseudomonas aeruginosa virulence proteins pseudolysin and protease IV impede cutaneous wound healing. Lab Invest 100, 1532–1550 (2020). https://doi.org/10.1038/s41374-020-00478-1. |
| [54] | Q. Wei, L.Z. Ma. Biofilm matrix and its regulation in Pseudomonas aeruginosa. Int J Mol Sci, 14 (2013), pp. 20983-21005. |
| [55] | R.M. Ostroff, A.I. Vasil, M.L. Vasil. Molecular comparison of a nonhemolytic and a hemolytic phospholipase C from Pseudomonas aeruginosa J Bacteriol, 172 (1990), pp. 5915-5923. |
| [56] | Rossolini GM, Mantengoli E, Docquier JD, Musmanno RA, Coratza G. Epidemiology of infections caused by multiresistant gram-negatives: ESBLs, MBLs, pan resistant strains. New Microbiol. 2007;30(3):332–9. |
| [57] | S. Ingle., K. Mahajan, B.S. Kumar, S. Singh, R.K. Agrawal, R., (2013). Verma Over-expression, and immunogenicity of outer membrane protein L (OprL) of Pseudomonas aeruginosa Proc Natl Acad Sci India Sec B Biol Sci (2015), pp. 1-7. |
| [58] | Sabharwal, N., Dhall, S., Chhibber, S., & Harjai, K. (2014). Molecular detection of virulence genes as markers in Pseudomonas aeruginosa isolated from urinary tract infections. International journal of molecular epidemiology and genetics, 5(3), 125–134. |
| [59] | Sadikot, R. T., Blackwell, T. S., Christman, J. W., & Prince, A. S. (2005). Pathogen-host interactions in Pseudomonas aeruginosa pneumonia. American journal of respiratory and critical care medicine, 171(11), 1209–1223. https://doi.org/10.1164/rccm.200408-1044SO. |
| [60] | Sanchez, C.J., Mende, K., Beckius, M.L. et al. Biofilm formation by clinical isolates and the implications in chronic infections. BMC Infect Dis 13, 47 (2013). https://doi.org/10.1186/1471-2334-13-47. |
| [61] | Saxena, S., Banerjee, G., Garg, R., & Singh, M. (2014). Comparative Study of Biofilm Formation in Pseudomonas aeruginosa Isolates from Patients of Lower Respiratory Tract Infection. Journal of clinical and diagnostic research: JCDR, 8(5), DC09–DC11. https://doi.org/10.7860/JCDR/2014/7808.4330. |
| [62] | Serghini MA, Ritzenthaler C, Pinck L (1989) A rapid and efficient 'miniprep' for isolation of plasmid DNA. Nucleic Acids Res 17: 3604. |
| [63] | Shaan L. Gellatly, Robert E.W. Hancock, Pseudomonas aeruginosa: new insights into pathogenesis and host defenses, Pathogens and Disease, Volume 67, Issue 3, April 2013, Pages 159–173, https://doi.org/10.1111/2049-632X.12033. |
| [64] | Shahcheraghi F, Feizabadi MM, Yamin V, Abiri R, Abedian Z. Serovar determination, drug resistance patterns and plasmid profiles of Pseudomonas aeruginosa isolated from burn patients at two hospitals of Tehran (IRAN). Burns. 2003; 29(6): 547–51. |
| [65] | Shaver, C. M., & Hauser, A. R. (2004). Relative contributions of Pseudomonas aeruginosa ExoU, ExoS, and ExoT to virulence in the lung. Infection and immunity, 72(12), 6969–6977. https://doi.org/10.1128/IAI.72.12.6969-6977.2004. |
| [66] | Singh, S., Singh, S. K., Chowdhury, I., & Singh, R. (2017). Understanding the Mechanism of Bacterial Biofilms Resistance to Antimicrobial Agents. The open microbiology journal, 11, 53–62. https://doi.org/10.2174/1874285801711010053. |
| [67] | Sousa, A. M., & Pereira, M. O. (2014). Pseudomonas aeruginosa Diversification during Infection Development in Cystic Fibrosis Lungs-A Review. Pathogens (Basel, Switzerland), 3(3), 680–703. https://doi.org/10.3390/pathogens3030680. |
| [68] | Tateda K, Comte R, Pechere JC, Kohler T, Yamaguchi K, Van Delden C., (2001). Azithromycin inhibits quorum sensing in Pseudomonas aeruginosa. Antimicrobial Agents Chemother; 45:1930-1933. |
| [69] | Tavajjohi Z, Moniri R, Khorshidi A. Detection and characterization of multidrug resistance and extended-spectrum-beta-lactamase-producing (ESBLS) Pseudomonas aeruginosa isolates in teaching hospital. African J Microbiol Res. 2011; 5(20): 3223–8. |
| [70] | Tsegaye Alemayehu, (2020). Prevalence of multidrug-resistant bacteria in Ethiopia: a systematic review and meta-analysis, Journal of Global Antimicrobial Resistance Volume 26, September 2021, Pages 133-139. https://doi.org/10.1016/j.jgar.2021.05.017. |
| [71] | W.H. Sperber, M.P. Doyle (eds.), Compendium of the Microbiological Spoilage 135. |
| [72] | Waheed Ullah, Muhammad Qasim, Hazir Rahman, Yan Jie, Noor Muhammad, Beta-lactamase-producing Pseudomonas aeruginosa: Phenotypic characteristics and molecular identification of virulence genes, Journal of the Chinese Medical Association, Volume 80, Issue 3, 2017, Pages 173-177, ISSN 1726-4901. |
| [73] | World Health Organization (WHO, 2007) www.who.int/whr/2007/whr07_en.pdf. |
| [74] | Zhang, S., Yang, G., Ye, Q., Wu, Q., Zhang, J., & Huang, Y. (2018). Phenotypic and Genotypic Characterization of Klebsiella pneumoniae Isolated from Retail Foods in China. Frontiers in microbiology, 9, 289. https://doi.org/10.3389/fmicb.2018.00289. |
| [75] | Zhao, X., Yu, Z., & Ding, T. (2020). Quorum-Sensing Regulation of Antimicrobial Resistance in Bacteria. Microorganisms, 8(3), 425. https://doi.org/10.3390/microorganisms8030425. |





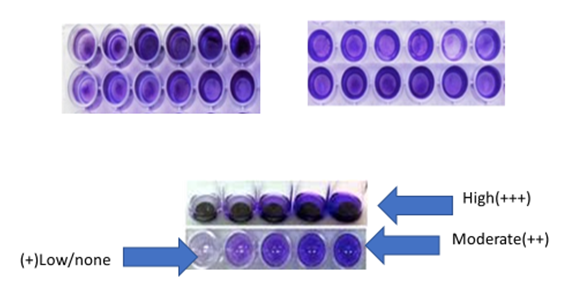
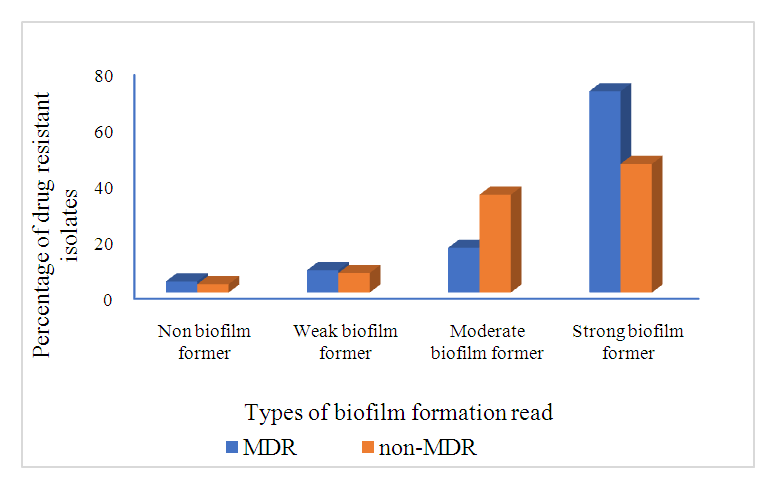
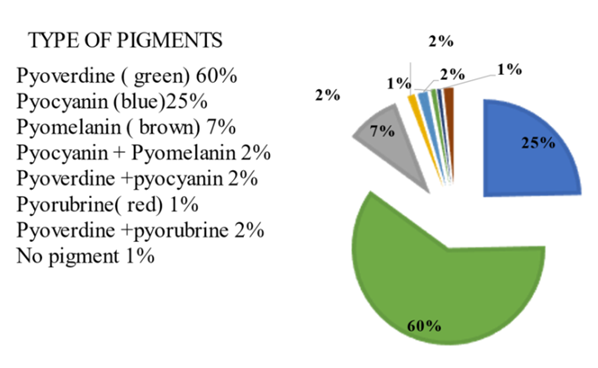
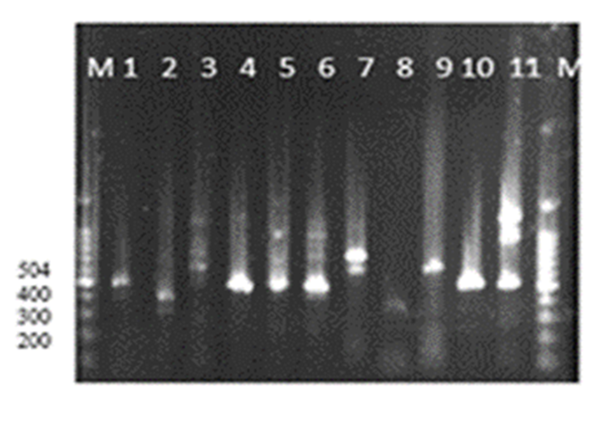
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML