| [1] | Huang Y.T., Hsueh P.R. (2008) Antimicrobial drug resistance in Taiwan. Int. J. Antimicrob. Agents; 32: S174–S178. |
| [2] | Pitout J.D. (2010) Infections with extended-spectrum beta-lactamase-producing Enterobacteriaceae: changing epidemiology and drug treatment choices. Drugs. 2010; 70: 313–333. |
| [3] | Nordmann P., Naas T., Poirel L. (2011) Global spread of carbapenemase-producing Enterobacteriaceae. Emerg. Infect. Dis. 17: 1791–1798. |
| [4] | Ørjan Samuelsen, Umaer Naseer, Nabil Karah, Paul Christoffer Lindemann, Anita Kanestrøm, Truls M. Leegaard, Arnfinn Sundsfjord, Identification of Enterobacteriaceae isolates with OXA-48 and coproduction of OXA-181 and NDM-1 in Norway, Journal of Antimicrobial Chemotherapy, Volume 68, Issue 7, July 2013, Pages 1682–1685, https://doi.org/10.1093/jac/dkt058. |
| [5] | Lee C-R., Lee J.H., Park K.S., Kim Y.B., Jeong B.C., Lee S.H. (2016) Global dissemination of carbapenemase-producing Klebsiella pneumoniae: epidemiology, genetic context, treatment options, and detection methods. Front. Microbiol. 7: 895. |
| [6] | Pournaras S., Kristo I., Vrioni G., Ikonomidis A., Poulou A., Petropoulou D., et al. (2010) Characteristics of meropenem heteroresistance in Klebsiella pneumoniae carbapenemase (KPC)-producing clinical isolates of K. pneumoniae. J. Clin. Microbiol. 48: 2601–2604. |
| [7] | Yigit H., Queenan A.M., Anderson G.J., Domenech-Sanchez A., Biddle J.W., Steward C.D., et al. (2001) Novel carbapenem-hydrolyzing beta-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 45: 1151–1161. |
| [8] | Yong D., Toleman M.A., Giske C.G., Cho H.S., Sundman K., Lee K., et al. (2009) Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 53:5046–5054. |
| [9] | Palacios, M., Miner, T. A., Frederick, D. R., Sepulveda, V. E., Quinn, J. D., Walker, K. A., & Miller, V. L. (2018). Identification of Two Regulators of Virulence That Are Conserved in Klebsiella pneumoniae Classical and Hypervirulent Strains. mBio, 9(4), e01443-18. https://doi.org/10.1128/mBio.01443-18. |
| [10] | Walther-Rasmussen J., Høiby N. (2006) OXA-type carbapenemases. J. Antimicrob. Chemother. 2006; 57(3): 373–383. |
| [11] | Kaczmarek F.M., Dib-Hajj F., Shang W., Gootz T.D. (2006) High-level carbapenem resistance in a Klebsiella pneumoniae clinical isolate is due to the combination of blaACT-1 β-lactamase production, porin OmpK35/36 insertional inactivation, and down-regulation of the phosphate transport porin PhoE. Antimicrobes. Agents Chemother. 50: 3396–3406. |
| [12] | Wang X.D., Cai J.C., Zhou H.W., Zhang R., Chen G.X. (2009) Reduced susceptibility to carbapenems in Klebsiella pneumoniae clinical isolates associated with plasmid-mediated beta-lactamase production and OmpK36 porin deficiency. J. Med. Microbiol. 58: 1196–1202. |
| [13] | Zhang, S., Yang, G., Ye, Q., Wu, Q., Zhang, J., & Huang, Y. (2018). Phenotypic and Genotypic Characterization of Klebsiella pneumoniae Isolated from Retail Foods in China. Frontiers in microbiology, 9, 289. https://doi.org/10.3389/fmicb.2018.00289. |
| [14] | Pfeifer Y, Schlatterer K, Engelmann E, Schiller RA, Frangenberg HR, Stiewe D, Holfelder M, Witte W, Nordmann P, Poirel L (2012) Emergence of OXA-48-type carbapenemase-producing Enterobac-teriaceae in German hospitals. Antimicrobe Agents Chemother56: 2125–2128. http://dx.doi.org/10.1128/AAC.05315-11. |
| [15] | Azap Ö, Otlu B, Yeşilkaya A, Yakupoğulları Y (2013) Detection of OXA-48 like carbapenemase-producing Klebsiella pneumoniae in a tertiary care center in Turkey: molecular characterization and epi-demiology. Balkan Med J30: 259–260. http://dx.doi.org/10.5152/balkanmedj.2013.7499. |
| [16] | Balm MND, La MV, Krishnan P, Jureen R, Lin RTP and Teo JWP (2013) Emergence of Klebsiella pneumoniae coproducing NDM-type and OXA-181 carbapenemases.Clin Microbiol Infec19: E421–E423. http://dx.doi.org/10.1111/1469-0691.12247. |
| [17] | Doi Y, O’Hara JA, Lando JF, Querry AM, Townsend BM, Pasculle AW, Muto CA (2014). Co-Production of NDM-1 and OXA-232 by Klebsiella pneumoniae. Emerg Infec Dis20:163–164. http://dx.doi.org/10.3201/eid2001.130904. |
| [18] | Shibl A, Al-Agamy M, Memish Z, Senok A, Khader SA, Assiri A (2013) The emergence of OXA-48- and NDM-1-positive Klebsiella pneumoniae in Riyadh, Saudi Arabia. Int J InfectDis17: e1130–e1133. http://dx.doi.org/10.1016/j.ijid.2013.06.016. |
| [19] | M. Saı¨dani, et.al., (2012) Emergence of carbapenem-resistant OXA-48 carbapenems-producing Enterobacteriaceae in Tunisia, Journal of Medical Microbiology, 61, 1746–1749. |
| [20] | Sunday B. et.al., (2016) Microbes in Irrigation Water and Fresh Vegetables: Potential Pathogenic Bacteria Assessment and Implications for Food Safety, https://doi.org/10.1177/1535676016652231 |
| [21] | Esra D. et.al. (2015) Klebsiella pneumoniae: Characteristics of carbapenem resistance and virulence factors. Vol. 62, No 4/2015. http://dx.doi.org/10.18388/abp.2015_1148 pp. 867-874. |
| [22] | Beceiro, A., Tomás, M., & Bou, G. (2013). Antimicrobial resistance and virulence: a successful or deleterious association in the bacterial world? Clinical microbiology reviews, 26(2), 185–230. https://doi.org/10.1128/CMR.00059-12. |
| [23] | Gupta N., Limbago B.M., Patel J.B., Kallen A.J. (2011) Carbapenem-resistant Enterobacteriaceae: epidemiology and prevention. Clin. Infect. Dis. 53:60–67. |
| [24] | Poirel L., Héritier C., Tolün V., Nordmann P. (2004) Emergence of oxacillinase-mediated resistance to imipenem in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 48: 15–22. |
| [25] | Shi W., Li K., Ji Y., Jiang Q., Wang Y., Shi M., et al. (2013) Carbapenem and cefoxitin resistance of Klebsiella pneumoniae strains associated with porin OmpK36 loss and DHA-1 β-lactamase production. Braz. J. Microbiol. 44: 435–442. |
| [26] | Steinmann J., Kaase M., Gatermann S., Popp W., Steinmann E., Damman M., et al. (2011) Outbreak due to a Klebsiella pneumoniae strain harbouring KPC-2 and VIM-1 in a German university hospital, July 2010 to January 2011. Euro Surveill. 16:19944. |
| [27] | Sahin K., Tekin A., Ozdas S., Akin D., Yapislar H., Dilek A.R., et al. (2015) Evaluation of carbapenem resistance using phenotypic and genotypic techniques in Enterobacteriaceae isolates. Ann. Clin. Microbiol. Antimicrob. 14:44. |
| [28] | Nordmann P., Poirel L. (2014) The difficult-to-control spread of carbapenemase producers among Enterobacteriaceae worldwide. Clin. Microbiol. Infect. 20:821–830. |
| [29] | Steinmann J., Kaase M., Gatermann S., Popp W., Steinmann E., Damman M., et al. (2011). Outbreak due to a Klebsiella pneumoniae strain harbouring KPC-2 and VIM-1 in a German university hospital, July 2010 to January 2011. Euro Surveill. 16:19944. |
| [30] | Perilli M., Bottoni C., Grimaldi A., Segatore B., Celenza G., Mariani M., et al. (2013) Carbapenem-resistant Klebsiella pneumoniae harbouring blaKPC-3 and blaVIM-2 from central Italy. Diagn. Microbiol. Infect. Dis. 75:218–221. |
| [31] | Rojas L.J., Mojica M.F., Blanco V.M., Correa A., Montealegre M.C., De La Cadena E., et al. Emergence of Klebsiella pneumoniae coharboring KPC and VIM carbapenemases in Colombia. Antimicrob. Agents Chemother. 2013; 57:1101–1102. |
| [32] | Brink, A. J., Coetzee, J., Corcoran, C., Clay, C. G., Hari-Makkan, D., Jacobson, R. K., Richards, G. A., Feldman, C., Nutt, L., van Greune, J., Deetlefs, J. D., Swart, K., Devenish, L., Poirel, L., & Nordmann, P. (2013). Emergence of OXA-48 and OXA-181 carbapenemases among Enterobacteriaceae in South Africa and evidence of in vivo selection of colistin resistance as a consequence of selective decontamination of the gastrointestinal tract. Journal of clinical microbiology, 51(1), 369–372. https://doi.org/10.1128/JCM.02234-12. |
| [33] | Effah CY, Sun T, Liu S, Wu Y. Klebsiella pneumoniae: an increasing threat to public health. Ann Clin Microbiol Antimicrob. 2020 Jan 9; 19(1): 1. doi: 10.1186/s12941-019-0343-8. PMID: 31918737; PMCID: PMC7050612. |
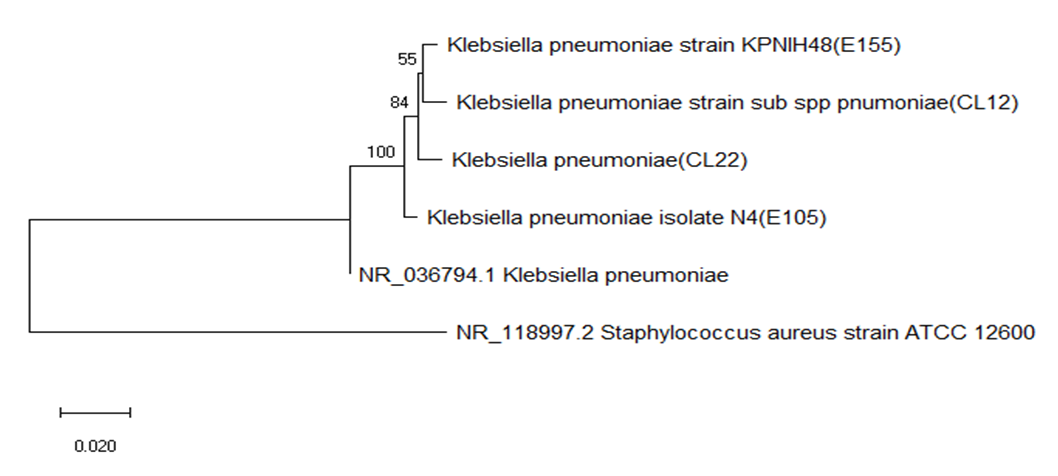
| [34] | Richard D. et.al., 1994. A Phylogenetic Tree of 16S rRNA Sequences from Sulfate Reducing Bacteria in a Sandy Marine Sediment. Applied and environmental Microbiology, Sept. 1994, p. 3437-3439, Vol. 60, No. |
| [35] | Hammoudi Halat D, Ayoub Moubareck C. The Current Burden of Carbapenemases: Review of Significant Properties and Dissemination among Gram-Negative Bacteria. Antibiotics (Basel). 2020 Apr 16; 9(4): 186. doi: 10.3390/antibiotics9040186. PMID: 32316342; PMCID: PMC7235769. |
| [36] | Codjoe, F. S., & Donkor, E. S. (2017). Carbapenem Resistance: A Review. Medical sciences (Basel, Switzerland), 6(1), 1. https://doi.org/10.3390/medsci6010001. |
| [37] | Queenan AM, Bush K. Carbapenemases: the versatile beta-lactamases. Clin Microbiol Rev. 2007 Jul; 20(3): 440-58, table of contents. doi: 10.1128/CMR.00001-07. PMID: 17630334; PMCID: PMC1932750. |
| [38] | Chavez, M. V., Caicedo, L. D., & Castillo, J. E. (2019). Occurrence of β-Lactamase-Producing Gram-Negative Bacterial Isolates in Water Sources in Cali City, Colombia. International journal of microbiology, 2019, 1375060. https://doi.org/10.1155/2019/1375060. |
| [39] | Bush K. Past and Present Perspectives on β-Lactamases. Antimicrob Agents Chemother. 2018 Sep 24; 62(10): 01076-18. doi: 10.1128/AAC.01076-18. PMID: 30061284; PMCID: PMC6153792. |
| [40] | Sheu, C. C., Chang, Y. T., Lin, S. Y., Chen, Y. H., & Hsueh, P. R. (2019). Infections Caused by Carbapenem-Resistant Enterobacteriaceae: An Update on Therapeutic Options. Frontiers in microbiology, 10, 80. https://doi.org/10.3389/fmicb.2019.00080. |
| [41] | Akinde1, S. B., Abiodun A. Sunday, A. A., Folasade M. Adeyemi, F. M., Iyabobola B. Fakayode1, I. B, Odunola O. Oluwajide, O.O., Adetoun A. Adebunmi1, A. A., Julius K. Oloke, J. K., and Clement O. Adebooye. C.O. (2016). Microbes in Irrigation Water and Fresh Vegetables: Potential Pathogenic Bacteria Assessment and Implications for Food Safety, Applied Biosafety: Journal of ABSA International 2016, Vol. 21(2) 89-97. |
| [42] | Venkatesan, M. M., Goldberg, M. B., Rose, D. J., Grotbeck, E. J., Burland, V., & Blattner, F. R. (2001). Complete DNA sequence and analysis of the large virulence plasmid of Shigella flexneri. Infection and immunity, 69(5), 3271–3285. https://doi.org/10.1128/IAI.69.5.3271-3285.2001. |
| [43] | Clarridge J. E., 3rd (2004). Impact of 16S rRNA gene sequence analysis for identification of bacteria on clinical microbiology and infectious diseases. Clinical microbiology reviews, 17(4), 840–862. https://doi.org/10.1128/CMR.17.4.840-862.2004. |
| [44] | Janda, J. M., & Abbott, S. L. (2007). 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: pluses, perils, and pitfalls. Journal of clinical microbiology, 45(9), 2761–2764. https://doi.org/10.1128/JCM.01228-07. |
| [45] | Karlowsky, J. A., Jones M. E., Thornsberry C., Friedland I. R., and Sahm D. F. (2003) Trends in antimicrobial susceptibilities among Enterobacteriaceae isolated from hospitalized patients in the United States from 1998 to 2001. Antimicrob. Agents Chemother. 47: 1672– 1680. |
| [46] | Siu, L. K, Fung C. P., Chang F. Y., Lee N., Yeh K. M., Koh T. H., and Ip M. (2011) Molecular typing and virulence analysis of serotype K1 Klebsiella pneumoniae strains isolated from liver abscess patients and stool samples from noninfectious subjects in Hong Kong, Singapore, and Taiwan. J. Clin. Microbiol. 49 (11): 3761– 3765. |
| [47] | Fung, C. P., Lin Y. T., Lin J. C., Chen T. L., Yeh K. M., Chang F. Y., Chuang H. C., Wu H. S., Tseng C. P., and Siu L. K. (2012) Klebsiella pneumoniae in gastrointestinal tract and pyogenic liver abscess. Emerg. Infect. Dis. 18 (8): 1322– 1325. |
| [48] | Lübbert, C., Wiegand J., and Karlas T. (2014) Therapy of liver abscess. Gastrointest. Med. Surg. 30: 334– 341. |
| [49] | Rahimian, J., Wilson T., Oram V., and Holzman R. S. (2004) Pyogenic liver abscess: recent trends in etiology and mortality. Clin. Infect. Dis. 39: 1654– 1659. |
| [50] | Siu, L. K, Yeh K. M., Lin J. C., Fung C. P., and Chang F. Y. (2012) Klebsiella pneumoniae liver abscess: a new invasive syndrome. Lancet Infect. Dis. 12: 881– 887. |
| [51] | Arnold, R. S., Thom K. A., Sharma S., Philips M., Johnson J. K., and Morgan D. J. (2011) Emergence of Klebsiella pneumoniae carbapenemase (KPC)–producing bacteria. South Med. J. 104 (1): 40– 45. |
| [52] | Paterson, D. L., and Bonomo R. A. (2005) Extended-spectrum B-lactamases: a clinical update. Clin. Microbiol. Rev. 18 (4): 657– 686. |
| [53] | Pitout, J. D. D., Nordmann P., and Poirel L. (2015) Carbapenemase-producing Klebsiella pneumoniae a key pathogen set for global nosocomial dominance. Antimicrob. Agents Chemother. 59 (10): 5873– 5884. |
| [54] | Kidd, T. J., Mills G., Sa-Pessoa J., Dumigan A., Frank C. G., Insua J. L., Ingram R., Hobley L., and Bengoechea J. A. (2017) A Klebsiella pneumoniae antibiotic resistance mechanism that subdues host defences and promotes virulence. EMBO Mol. Med. 9 (4): 430– 447. |
| [55] | Shroeder, M., Brooks B. D., and Brooks A. E. (2017) The complex relationship between virulence and antibiotic resistance. Genes 8 (39): 1– 23. |
| [56] | Calbo, E., Freixas N., Xercavins M., Riera M., Nicolás C., Monistrol O., Solé Mdel M., Sala M. R., Vila J., and Garau J. (2011) Foodborne nosocomial outbreak of SHV1 and CTX-M-15–producing Klebsiella pneumoniae: epidemiology and control. Clin. Infect. Dis. 52 (6): 743– 749. |
| [57] | Davis, G. S., Waits K., Nordstrom L., Weaver B., Aziz M., Gauld L., Grande H., Bigler R., Horwinski J., Porter S., Stegger M., Johnson J. R., Liu C. M., and Price L. B. (2015) Intermingled Klebsiella pneumoniae populations between retail meats and human urinary tract infections. Clin. Infect. Dis. 61 (6): 892– 899. |
| [58] | Falomir, M. P., Rico H., and Gozalbo D. (2013) Enterobacter and Klebsiella species isolated from fresh vegetables marketed in Valencia (Spain) and their clinically relevant resistances to chemotherapeutic agents. Foodborne Pathog. Dis. 10: 1002– 1007. |
| [59] | Ghenghesh, K. S., Belhaj K., El-Amin W. B., El-Nefathi S. E., and Zalmum A. (2004) Microbiological quality of fruit juices sold in Tripoli-Libya. Food Control 16 (10): 855– 858. |
| [60] | Guo, Y., Zhou H., Qin L., Pang Z., Qin T., Ren H., Pan Z., and Zhou J. (2016) Frequency, antimicrobial resistance, and genetic diversity of Klebsiella pneumoniae in food samples. PLoS ONE 11 (4): e0153561. |
| [61] | Kim, S. H., Wei C. I., Tzou Y. M., and An H. (2005) Multidrug-resistant Klebsiella pneumoniae isolated from farm environments and retail products in Oklahoma. J. Food Prot. 68 (10): 2022– 2029. |
| [62] | Puspanadan, S., Afsah-Hejri L., Loo Y. Y., Nillian E., Kuan C. H., Goh S. G., Chang W. S., Lye Y. L., John Y. H. T., Rukayadi Y., Yoshitsugu N., Nishibuchi M., and Son R. (2012) Detection of Klebsiella pneumoniae in raw vegetables using most probable number-polymerase chain reaction (MPN-PCR). Int. Food Res. J. 19 (4): 1757– 1762. |
| [63] | Wu, H., Liu B. G., Liu J. H., Pan Y. S., Yuan L., and Hu G. Z. (2012) Phenotypic and molecular characterization of CTX-M-14 extended-spectrum β-lactamase and plasmid-mediated ACT-like AmpC β-lactamase produced by Klebsiella pneumoniae isolates from chickens in Henan Province, China. Genet. Mol. Res. 11 (3): 3357– 3364. |
| [64] | Haryani, Y., Noorzaleha A. S., Fatimah A. B., Noorjahan B. A., Patrick G. B., Shamsinar A. T., Laila R. A. S., and Son R. (2007) Incidence of Klebsiella pneumoniae in street foods sold in Malaysia and their characterization by antibiotic resistance, plasmid profiling, and RAPD-PCR analysis. Food Control 18: 847– 853. |
| [65] | Sabota, J. M., Hoppes W. L., Ziegler J. R., DuPont H., Mathewson J., and Rutecki G. W. (1998) A new variant of food poisoning: enteroinvasive Klebsiella pneumoniae and Escherichia coli sepsis from a contaminated hamburger. Am. J. Gastroenterol. 93: 118– 119. |
| [66] | Tooke, C. L., Hinchliffe, P., Bragginton, E. C., Colenso, C. K., Hirvonen, V., Takebayashi, Y., & Spencer, J. (2019) β-Lactamases and β-Lactamase Inhibitors in the 21st Century. Journal of molecular biology, 431(18), 3472–3500. https://doi.org/10.1016/j.jmb.2019.04.002. |
| [67] | Akoachere, F. T., Bertrand, F., and Joseph, M. N. (2018). Bacterial and parasitic contaminants of salad vegetables sold in markets in Fako Division, Cameroon and evaluation of hygiene and handling practices of vendors. BMC Research Notes, 11, 100. https://doi.org/10.1186/s13104-018-3175-2. |
| [68] | Asmaru, G., Samuel S. (2013). Microbial Spectrum of Fruit in Gondar town Markets, Northwestern Ethiopia, Journal of Microbiology Research, 3(1): 1-10. doi: 10.5923/j.microbiology.20130301.01. |
| [69] | Bruce, S. K., Schick, D. G., Tanaka, L., Jimenez, E. M., & Montgomerie, J. Z. (1981). Selective medium for isolation of Klebsiella pneumoniae. Journal of clinical microbiology, 13(6), 1114–1116. https://doi.org/10.1128/jcm.13.6.1114-1116.1981. |
| [70] | Bennett P. M. (2008). Plasmid encoded antibiotic resistance: acquisition and transfer of antibiotic resistance genes in bacteria. British journal of pharmacology, 153 Suppl 1 (Suppl 1), S347–S357. https://doi.org/10.1038/sj.bjp.0707607. |
| [71] | Bruce Alberts, Alexander Johnson, Julian Lewis, Martin Raff, Keith Roberts, and Peter Walter (2002) Molecular Biology of the Cell, 4th edition |
| [72] | Moges, F., Endris, M., Mulu, A., Tessema, B., Belyhun, Y., Shiferaw, Y., Huruy, K., Unakal, C., & Kassu, A. (2014). The growing challenges of antibacterial drug resistance in Ethiopia. Journal of global antimicrobial resistance, 2(3), 148–154. https://doi.org/10.1016/j.jgar.2014.02.004. |
| [73] | El Fertas-Aissani, R., Messai, Y., Alouache, S., & Bakour, R. (2013). Virulence profiles and antibiotic susceptibility patterns of Klebsiella pneumoniae strains isolated from different clinical specimens. Pathologie-biologie, 61(5), 209–216. https://doi.org/10.1016/j.patbio.2012.10.004. |
| [74] | Mishra, M., Panda, S., Barik, S., Sarkar, A., Singh, D. V., & Mohapatra, H. (2020). Antibiotic Resistance Profile, Outer Membrane Proteins, Virulence Factors and Genome Sequence Analysis Reveal Clinical Isolates of Enterobacter Are Potential Pathogens Compared to Environmental Isolates. Frontiers in cellular and infection microbiology, 10, 54. https://doi.org/10.3389/fcimb.2020.00054. |
| [75] | A. W. Bauer, M.D., W. M. M. Kirby, M.D., J. C. Sherris, M.D., M. Turck, M.D., Antibiotic Susceptibility Testing by a Standardized Single Disk Method, American Journal of Clinical Pathology, Volume 45, Issue 4_ts, April 1966, Pages 493–496, https://doi.org/10.1093/ajcp/45.4_ts.493. |
| [76] | Joseph Felsenstein, An Alternating Least Squares Approach to Inferring Phylogenies from Pairwise Distances, Systematic Biology, Volume 46, Issue 1, March 1997, Pages 101–111, https://doi.org/10.1093/sysbio/46.1.101. |
| [77] | Kuş, H., Arslan, U., Türk Dağı, H., & Fındık, D. (2017). Hastane enfeksiyonu etkeni Klebsiella pneumoniae izolatlarında çeşitli virülans faktörlerinin araştırılması [Investigation of various virulence factors of Klebsiella pneumoniae strains isolated from nosocomial infections]. Mikrobiyoloji bulteni, 51(4), 329–339. https://doi.org/10.5578/mb.59716. |
| [78] | Barguigua, A., El Otmani, F., Lakbakbi El Yaagoubi, F., Talmi, M., Zerouali, K., & Timinouni, M. (2013). First report of a Klebsiella pneumoniae strain coproducing NDM-1, VIM-1 and OXA-48 carbapenemases isolated in Morocco. APMIS: acta pathologica, microbiologica, et immunologica Scandinavica, 121(7), 675–677. https://doi.org/10.1111/apm.12034. |
| [79] | Contreras, D. A., Fitzwater, S. P., Nanayakkara, D. D., Schaenman, J., Aldrovandi, G. M., Garner, O. B., & Yang, S. (2020). Coinfections of Two Strains of NDM-1- and OXA-232-Coproducing Klebsiella pneumoniae in a Kidney Transplant Patient. Antimicrobial agents and chemotherapy, 64(4), e00948-19. https://doi.org/10.1128/AAC.00948-19. |
| [80] | P. Nordmann and L. Poirel1, (2014) The difficult-to-control spread of carbapenem’s producers among Enterobacteriaceae worldwide, 10.1111/1469-0691.12719. |



 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML