-
Paper Information
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
Journal of Microbiology Research
p-ISSN: 2166-5885 e-ISSN: 2166-5931
2021; 11(1): 8-20
doi:10.5923/j.microbiology.20211101.02
Received: Sep. 19, 2021; Accepted: Sep. 30, 2021; Published: Oct. 15, 2021

Prevalence of Carbapenem Resistant Acinetobacter Baumannii in Leafy Vegetable Samples and Clinical Sources from Gondar Northwest Ethiopia
Cherinet Yigrem1, Azanaw Amare2, Broderick Eribo3
1Department of Biology, Howard University, 400 Bryan St. NW Washington DC, USA
2Department of Microbiology, University of Gondar, Gondar, Ethiopia
3Department of Biology, Howard University, 415 College St. NW Washington DC, USA
Copyright © 2021 The Author(s). Published by Scientific & Academic Publishing.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/

Acinetobacter baumannii has been an important nosocomial pathogen causing infections in immunocompromised and hospitalized patients. Multiple antibiotic resistant A. baumannii isolates are spreading and causing outbreaks globally. The present study was done to assess the prevalence of antibiotic resistance pattern of A. baumannii isolated from leafy vegetable samples from different retail markets and clinical strains collected from Gondar University Hospital. Among the 13(48.1%) vegetables and 14(51.9%) clinical isolates, the highest Acinetobacter baumannii was detected in cauliflower 6/27 (22.2%). A. baumannii isolates harbored the highest prevalence of resistance against 23 antibiotics including the following carbapenem antibiotics, meropenem (97%), imipenem (99%), ertapenem (94%), doripeneme (96.6%). Comparison of antibiotic susceptibility rate between the vegetable and clinical isolates based on one-way A nova test (P=0.93>0.05) confirmed multiple antibiotic resistance (MDR). Phylogenetic tree determined the relationship of the vegetable isolates, and the clinical isolates were coherent each other. PCR analysis result of 9 representative isolates demonstrated 100% of them harbor resistance determinants (bla OXA23), (88.8%) of them harbor resistance determinants ompA, 100% harbor resistance determinants blaOXA51 and 100% harbor resistance determinants ISAab1. Furthermore, plasmid extraction analysis for 21 representative isolates confirmed that all the isolates harbor at least one plasmid.
Keywords: A. baumannii, Carbapenem resistant, Plasmid, Resistance genes, Virulence factors
Cite this paper: Cherinet Yigrem, Azanaw Amare, Broderick Eribo, Prevalence of Carbapenem Resistant Acinetobacter Baumannii in Leafy Vegetable Samples and Clinical Sources from Gondar Northwest Ethiopia, Journal of Microbiology Research, Vol. 11 No. 1, 2021, pp. 8-20. doi: 10.5923/j.microbiology.20211101.02.
Article Outline
1. Introduction
- Acinetobacter baumannii is an opportunistic pathogen, which has recently been successfully spreading worldwide nosocomial infections and causing outbreaks of hospital acquired infections, primarily due to its prominent ability to accumulate antibiotic resistant determinants [28,29,39,55,56]. However, despite hospital ecology being intensively studied, its ecology outside the hospital remains unclear or poorly defined impeding effective prevention of transmission [17,41]. A lot of researchers have a doubt that the survival of A. baumannii within the environment (in vegetables and water) could contribute to the transmission of the organism during outbreaks [45]. Prevalence of A. baumannii has been reported from hospital environments; however, only few studies have reported the presence of A. baumannii within the vegetable foods and water environment, meat of food animals and thus the environment where these meats come from [57]. These areas are a source of water and food and thus are often of transmission of this organism into hospital settings or may horizontally transfer resistance genes to other organisms [58]. Well established reports indicate that members of the genus Acinetobacter are ubiquitous but difficulty in unequivocally identify this organism has made the ubiquity in nature of A. baumannii a widespread misconception [27]. Therefore, the existence of an additional hospital reservoir of A. baumannii and thus the implication of this potential reservoir within the occurrence of certain infections due to this organism are still controversial [59]. Additionally, the prevalence and antimicrobial resistance of A. baumannii outside the hospital setting has been poorly investigated [11]. There seems to be endless influx of novel strains into the clinical setting with the potential of latest infectious features [17]. Several studies reported that infections caused by extra-hospital strains of A. baumannii are liable for community-acquired infections mainly in tropical and subtropical areas [2,12]. This means a source of this pathogen outside of the hospital and thus the importance of environmental isolates within the epidemiology of A. baumannii is of an excellent concern worldwide.A. baumannii is transformed into multiple antibiotic resistant bacteria, and its adaptation to the environment with antibiotic resistance has been reported in the past [60]. Carbapenems are kept because the last antibiotic choice for the treatment of severe infections, due to its extremely effective antibacterial activity and low toxicity, but the emergence of carbapenem resistance during A. baumannii has become a worldwide concern recently. Resistance A. baumannii strains to expanded spectrum β-lactams, cephalosporins and carbapenems has been rapidly growing over the years [61,62].Diverse sorts of β-lactam antibiotics, for example carbapenems, accommodate β-lactam rings in their structures and may be deactivated by β-lactamase enzymes. There are four molecular groups of β-lactamases, Ambler classes A through D, persistent with aminoalkanoic acid sequence identities. Molecularly classified, class A, C, and D (OXA enzymes) of β-lactamases accommodate a catalytically active serine remainder that split the lactam ring of antibiotics [63,64]. Class B of β-lactamases attribute completely different mechanism for enzyme activity, the reason might be it is a metallo-enzyme that needs zinc for its catalytic activity [65]. Carbapenem antibiotics, like imipenem and meropenem, have a tremendously broad spectrum of activity and are ready to resist hydrolysis by most of the β-lactamases, including extended-spectrum and derepressed class C chromosomal AmpC β-lactamases [66]. However, most of the metallo-β-lactamases and variety of sophistication A and D β-lactamases are ready to hydrolyze broad spectrum carbapenems, like imipenem and meropenem [18,46,67]. Thus, outbreaks of OXA-23-producing A. baumannii are reported from various regions of the planet [68]. It is generally believed that OXA-23 is liable for carbapenem antibiotic resistance. The OXA-23 carbapenems has been found in Acinetobacter from cattle, horses, and cats [13,14,32]. It has been also reported also the NDM-1 resistance genes were detected in Acinetobacter spp. from food animals (chicken and pig farms) in China [26,43]. However, knowledge about carbapenems producing A. baumannii of environmental origin (vegetables and water) remains very limited, making it difficult to assess its impact on public health. Although, it has been reported that, green leafy samples had high microbial load and multiple antibiotic resistant pathogens in Ethiopia, the cause of resistance is not known yet [69,70]. The current study aimed to report the prevalence of antibiotic resistance among different A. baumannii isolates their plasmid profile, determining the phylogenetic relationship by sequencing the 16srRNA and determining A. baumannii harboring carbapenems genes and possible virulence factors throughout a vegetable and clinical isolate in Gondar Ethiopia.
2. Materials and Methods
2.1. Description of the Study Area
- The study was conducted at University of North Gondar. North Gondar is found within the northwestern Ethiopia at a latitude of 120 36’ N, and longitude of 370 28’ E with an elevation of 2080 m. a. s. l. North Gondar town is found within the west of Northern part of Gondar administrative zone which is way from 747 Km from Addis Ababa within the northwest direction. According to the 2019 census the entire population of Gondar is 500,788 (97,625 males and 97,148 females). Gondar has mid altitude climate and a mean annual max temperature of 27°C and minimum temperature of 16°C. North Gondar city has twenty-three urban and 11 rural kebeles (Gondar Statistics office) [50].
2.2. Sample Collection
- A total of 110 leafy greens including cauliflower (n = 28), lettuce (n = 28), spinach (n = 27), and cabbage (n = 27) samples were randomly collected from four different retail markets in Gondar Ethiopia (May 2016-Aug 2016). All samples were collected with following proper aseptic technique. Samples were immediately transferred to the microbiology research facility of the Gondar University refrigerator in cooler with ice packs. All vegetable samples showed normal natural characters including odor, color, and consolidation.
2.3. Bacterial Isolation
- From each sample 11g of vegetable was homogenized in 99 ml of Luria-Bertani broth during a stomacher (Stomacher 400 Circulator, Seward, Norfolk, UK) for 1 minutes and was incubated overnight at 37°C with agitation. Out of the 12-18hrs incubated culture, 10 µl was streaked onto selective Chrom ID ESBL agar (bioMérieux, Marcy-l’Étoile, France), which doesn't inhibit the expansion of A. baumannii [71] and was incubated overnight at 37°C. All white colonies (presumptive A. baumannii) were transferred to tryptic soy agar (TSA) plates containing 5% sheep blood (BD, Franklin Lakes, NJ) and were incubated 18-24hrs at 37°C. All the necessary biochemical methods, including oxidase, citrate, urease, malonate consumption, oxidation and fermentation of sugars, motility, and indole production, were used to identify A. baumannii. Moreover, the genus Acinetobacter was identified by Gram staining, cell and colony morphology, positive catalase test, negative oxidase test and absence of true motility. Virulence factors and other characteristics of Acinetobacter baumannii was performed including glucose oxidation, gelatin liquefaction, beta hemolysis, biofilm formation, proteolytic activity, motility assay, growth at 37°C and 42°C, arginine hydrolysis and susceptibility to chloramphenicol. The colonies were further identified using analytical profile index API 20 [71].
2.4. Antibiotic Susceptibility
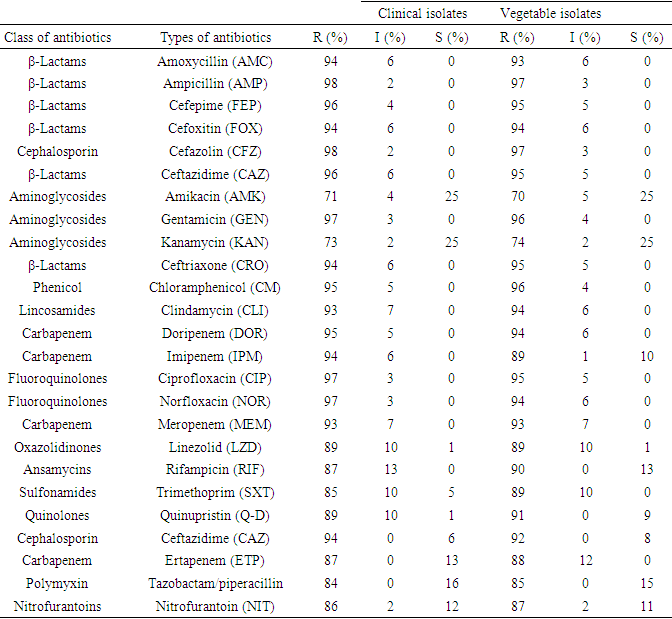
- Antimicrobial susceptibility of the A. baumannii isolates were performed using the simple disk diffusion technique. The experiment was done using the Kirby-Bauer method on Mueller Hinton agar (Merck, Darmstadt, Germany). The susceptibility of A. baumannii isolates were tested against 25 types of antibiotics with appropriate disks (Oxoid, Basingstoke, UK) were classified into 11 different groups [72]: amoxycillin(AMC, 30 μg), ampicillin (AMP, 10 μg), cefepime (FEP, 10 μg), Cefoxitin (FOX, 30 μg), Cefazolin (CFZ) (30 μg), ceftazidime (CAZ, 30 μg), amikacin (AK, 30 μg), gentamicin (CN, 10 μg), kanamycin (K, 30 μg), Cefaclor (CEC) (25μg), Cefotaxime (CTX) (30μg), clindamycin (DA, 2 μg), Doripenem (DOR) (30 μg), Imipenem (IPM) (30 μg), ciprofloxacin (CIP, 5 μg), Norfloxacin (NOR, 10 μg), Meropenem (MEM) (30μg), Linezolid (LZD, 30 μg), rifampicin (RD, 5 μg), Trimethoprim/sulphamethoxazole (SXT, 25 μg), Quinupristin/dalfopristin (QD, 15 μg), Ceftazidime (CAZ(30μg), Ertapenem(ETP) (30μg), Nalidixic acid (NAL) (30 μg) and Colistin (CST) (30 μg). The experiment was done according to the instruction of Clinical and Laboratory Standards Institute (CLSI, 2017). The reference strains A. baumannii ATCC 19606, Escherichia coli ATCC 25922 and Pseudomonas aeruginosa ATCC 27853 were used as quality controls.
2.5. Plasmid Profile and 16s rRNA Sequencing of Bacteria Isolates
- The plasmid identification of the isolates was performed by the OMEGA/miniprep method. 12-24 hours cultured bacteria isolate was prepared in 5 mL of nutrient broth. The broth culture was properly mixed by vertexing, and 1.5 mL was then transferred into a prelabeled Eppendorf tube. The tubes were then centrifuged for 2 minutes at 13 000 revolutions per minute (rpm) to harvest the bacterial cells. The remained supernatant was gently decanted, leaving about 100 μL of the broth culture, which was then vortexed at high speed until the bacterial cell pellet became completely suspended [73]. OMEGA solution (300 μL; 25mM Tris, 10mM EDTA, 0.1N NaOH, 0.5% SDS) was then added to lyse the bacterial cells. It was mixed by inversion 3 to 5 times until the solution became slimy, after which 150 μL of 3.0M sodium acetate (pH 5.2) was added and again vortexed for about 10 seconds. It was further centrifuged at 13 000 rpm for 5 minutes to pellet out cellular debris and chromosomal DNA. The remained supernatant was then transferred into another labeled 1.5mL Eppendorf tube, and 900 μL of cold absolute ethanol was added. The solution was centrifuged at 13 000 rpm for 10 minutes. The supernatant was discarded, and the white pellet containing the plasmid DNA was rinsed twice with 1000 μL of 70% ethanol [74]. The final pellet was then air-dried, and 40 μL of TE buffer (10mM Tris, 1mM Na2EDTA) was added to resuspend the pellet. Electrophoresis of the extracted plasmid DNA was carried out on 1.0% agarose gel and run for 3 hours at 200 V. It was visualized with ethidium bromide via the ultraviolet transilluminator. Representative isolates were sent to the GENWIZ for Sequencing of 16S rRNA gene [75,76].
2.6. Primers and PCR Confirmation of Resistance Genes
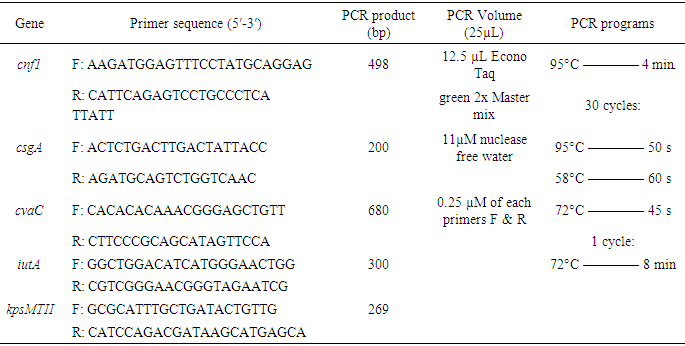
- All confirmed A. baumannii isolates were grown overnight in lactose broth at 37°C. Genomic DNA was extracted using a commercial Universal DNA Extraction Kit (Sangon Biotech, Shanghai, China) according to the manufacturer's instructions. Confirmation of A. baumannii isolates was performed by PCR as previously described. The primer sequences and amplicon size are shown in table 1 [77].Colony PCR was performed to detect all selected resistance and virulence genes using protocols listed in table 1. The PCR mixture will be obtained from EconoTaq PLUS and EconoTaq PLUS GREEN 2X Master Mixes, and all reactions were conducted under proper conditions: initial denaturation at 94°C for 4 minutes, followed by 35 cycles of 94°C for 45 seconds, polymerization at 72°C for 1 minute, and final extension for 5 minutes and 1 minute for annealing. Amplified products of all virulence factors will be further analyzed using ethidium bromide-mixed agarose gel (1.8%) under a UV Tran illuminator. Resistant genes will be determined from the isolates based on protocols listed in table 1 using standard primers by performing colony PCR as depicted in table 2 [78,79].
|
|
|
2.7. Biofilm Determination
- The biofilm formation capability of the bacterial isolates was determined using the microtiter-plate assay. After preparation of bacterial suspensions in trypticase soy broth (TSB) (Merck, Darmstadt, Germany) with an OD600 of 0.1, suspensions were inoculated in 96-well polystyrene microplates and incubated at 37°C for twenty-four h. Microplates were washed thrice with 1× phosphate buffered saline (PBS). These were then fixed using absolute methanol and stained using 1% gentian violet. The optical absorbance was recorded at 570 nm and results were analyzed and interpreted [80,81].
2.8. Motility Assay of Acinetobacter Baumannii
- Surface motility was detected based on a standard procedure (Vijayakumar, S. et al., 2016). One colony of A. baumannii was grown in Luria-Bertani (LB) (Merck) broth at 37°C for 24hrs. The bacterial suspension turbidity was adjusted to 0.5 McFarland (108 CFU/mL) then 10 µL of the suspension were cultured on LB agar plates of 0.3%. These plates were incubated at 37°C for twenty-four hrs. and therefore the growth area (cm2) was calculated. The bacterial motility was categorized supported the standards described previously.
2.9. Hemolysis Assay and Proteolytic Activity in Acinetobacter baumannii
- To identify hemolytic activity, one colony from A. baumannii was dissolved in LB broth (Merck) and incubated at 37°C for twenty-four h. This suspension was adjusted to 0.5 McFarland and 10 µL of the suspension were cultured on 5% agar (BA) (Merck) plates. These were then incubated at 37°C for one week. The proteolytic activity of A. baumannii was assessed supported previous reports. Briefly, one bacterial colony was suspended in TSB and incubated at 37°C for twenty-four h with agitation. The bacterial suspension (turbidity of 0.5 McFarland) was centrifuged, and therefore the supernatant was filter (0.22 µ) sterilized. Then, 0.5 mL of the bacterial supernatant were incubated with 0.5 mL of azoalbumin solution (1 mg/mL) (Merck) at 37°C for 24hrs with agitation (Phillips p, et al., 1984). After incubation, trichloracetic acid (13%) (Merck) was added to tubes and incubated at -20°C for 30 min (Phillips p, et al., 1984). The procedure was followed by centrifugation (1300g) for 10 min and therefore the OD440 of the resulting supernatants was measured [82].
2.10. Statistical Analysis
- To assess differences in aerobic mesophilic bacterial count, total coliform count and fecal bacteria count among the vegetable samples, performed using the serial dilutions (log10 of cfu/gm) and the standard plat count formula (number of colonies times dilution factor divided by volume). Overall statistical analysis was done using the SPSS 21.0 statistical software (SPSS Inc., Chicago, IL, USA). Chi-square test and Fisher's exact two‐tailed test were used to assess any significant relationship between the prevalence of A. baumannii isolates and their antibiotic resistance pattern. A value of P < .05 was considered as statistically significant level. Differences in overall clinically significant bacterial distribution among the vegetable types (caulis flower, lettuce, spinach, cabbage) were analyzed using average (mean) and percentage. Positive correlations in antibiotic sensitivity test between the bacterial isolates of the vegetable samples and clinical sources were assessed employing a per mutational multivariate ANOVA test (PERMANOVA). ANOVA tests were also wont to test for the resistance genes among the bacterial isolates. Significant differences in bacterial population were observed across vegetable types using the nonparametric Kruskal-Wallis test and between environmental samples and clinical sources employing a t-test [83].
3. Results
3.1. Total Viable Count Total of Vegetable Samples
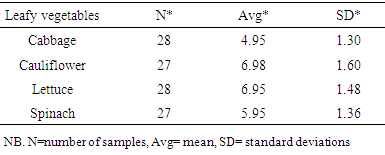
- The result of the total bacterial count, the average counts and standard deviation count for each leafy vegetable are calculated in table 4. The highest amount of mean count was recorded from cauliflower (6.98 cfu/g) and lettuce (6.95 cfu/g).
|
|
3.2. 16SrRNA Sequencing and Phylogenetic Relationship
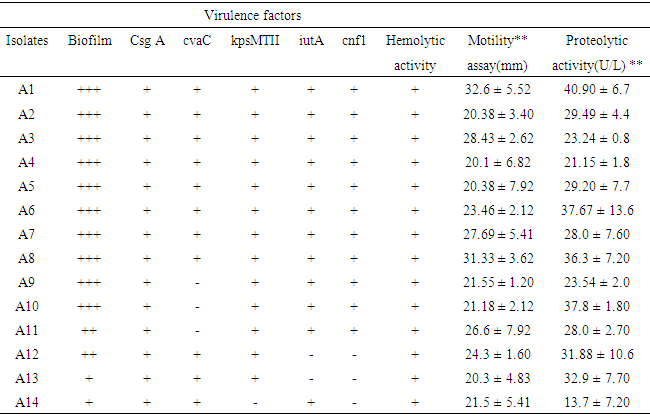
- All representative isolates were confirmed by 16SrRNA sequence. A. baumannii strain NF98 (MN871753), A. baumannii strain NF98, (MN871847), A. calcoaceticus strain BFHC18, (MN871921), Acinetobacter spp. clone PLB-D11, (MN871922), Acinetobacter spp. 121217-41_D05_SPAK1_518F, (MN871920), Acinetobacter spp. NF22, (MN872301), Acinetobacter sp. NF22, (MN872252). Sequences of vegetable isolates and clinical isolates were aligned together. Aligned sequences were compared with the same regions of 16S rRNAs from gene bank and other bacterial groups. Phylogenetic trees were constructed by the least square’s method (Richard D. et. al., 1994). Six sequences from both sources were considered to group within the gene bank sequenced species (Fig. 1). The lines represented by clinical isolated A. baumannii CL55 (MN871847) and vegetable isolated A. baumannii E33 (MN871753) shared 100% similarity with each other and shared 54% similarity in 16S rRNA sequence with sequence from the gene bank. All the vegetable and clinical Acinetobacter sequences were aligned and grouped together (figure 1).
 | Figure 1. Evolutionary relationship of 16s rRNA sequenced Acinetobacter spp., (NR-026206.1 A. baumannii strain is from NCBI) |
3.3. Plasmid Isolation and Profiling
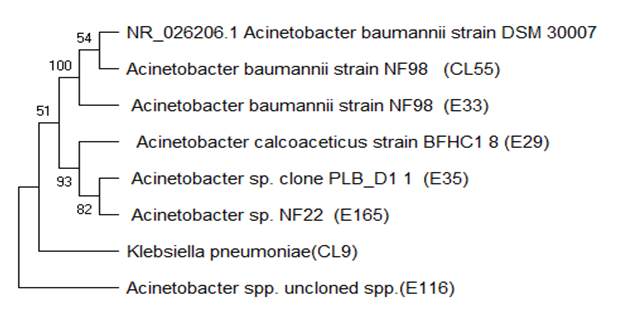
- Plasmid profile was carried out on the 21 of the 27 multiple antibiotic resistance Acinetobacter baumannii isolates. All the isolates 15(55.6%) were found to possess at least one plasmid bands. Plasmids were detected from eight of the clinical isolates (1-11), the other five of the plasmids detected from vegetable isolates (12-21) as depicted in figure 2 below.
 | Figure 2. Plasmid profile of Acinetobacter baumannii (a) Lane 1-11 clinical isolates, Lane 12-21 vegetable isolates |
3.4. Analyzing the PCR Result for Resistance Genes
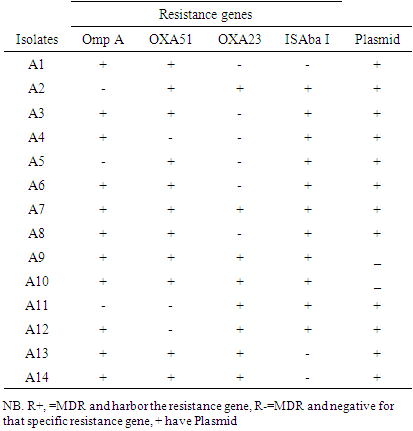
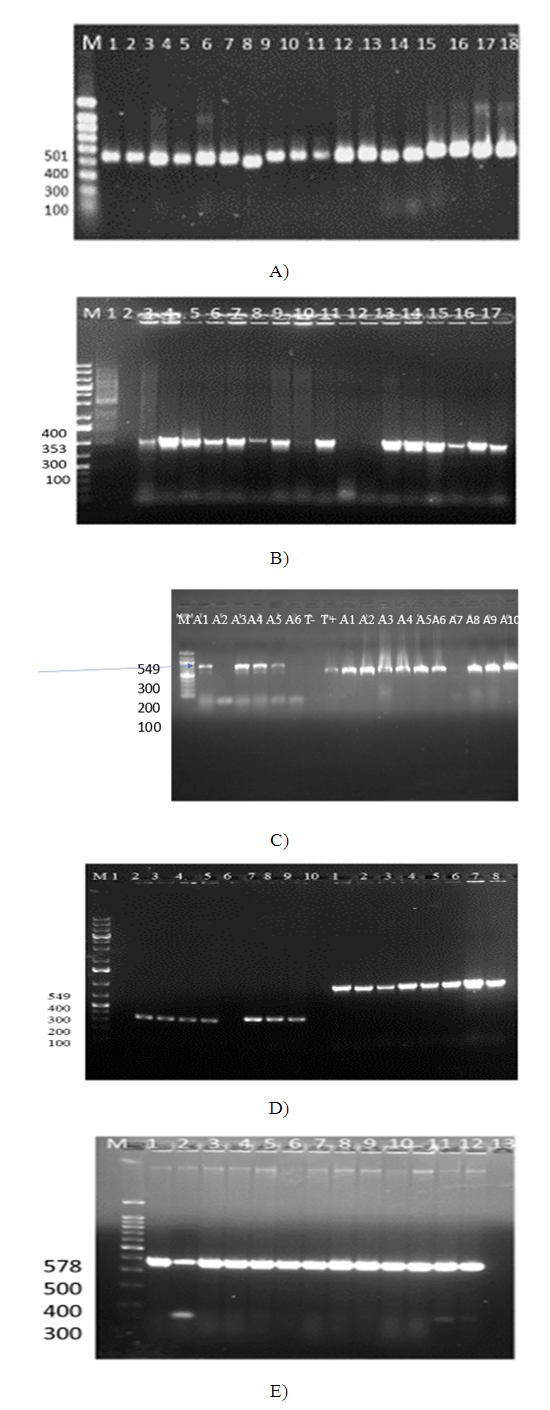
- Seven of the vegetable isolates and nine of the clinical isolates carried blaOXA-51. The blaOXA-23 was detected in 10(66.7%) vegetable isolates and 8(21.2%) clinical isolates. The ISAba I (549bp) was positive in 8 (14.6%) the clinical isolates. The omp A (578bp) resistance gene was detected in 4/10 of the colony PCR tested clinical isolates only. Out of the ten randomly selected vegetable isolates for colony PCR all of them lacked ISAba1 and ompA (figure 3. A, B, C, D, E) and table 6.
|
3.5. Analysis of Virulence Factors
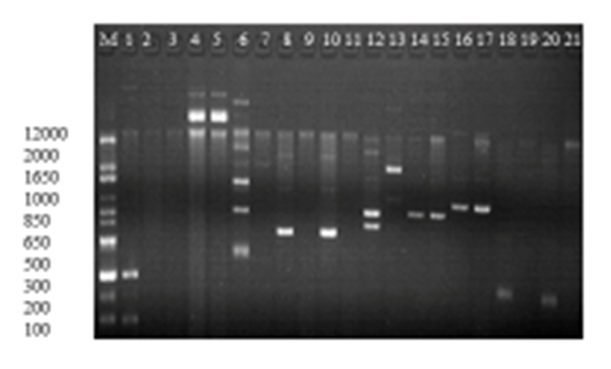
- For analysis of biofilm formation assay, OD570 values (mean ± standard deviation) of the negative control included 0.012±0.014. These values ranged between 0.013-0.024 in 3 isolates interpreted as negative (-), 0.05-0.06 in 2 isolates interpreted as weak (+), 0.09-0.13 in 10 isolates interpreted as moderate (++) and 0.17-0.24 in 12 isolates interpreted as strong (+++) biofilm producers (table 7). Biofilm producing A. baumannii showed (96%, 95%), (94%, 94%), (71%, 70%), (73%, 74%), (94%, 89%) and (93%, 93%) resistance to cefepime, cefoxitin, amikacin, kanamycin, imipenem and meropenem, in clinical and vegetable isolates respectively. Resistance to aminoglycosides, beta lactamase and carbapenem resistant were comparatively higher among biofilm producing A. baumannii than nonproducer (Statistically significant; P < 0.001). MDR were comparatively higher among biofilm producing than nonproducer (statistically significant; P < 0.001). Relationship between MDR pattern of biofilm producing and nonproducing A. baumannii are shown in figure 4 below.
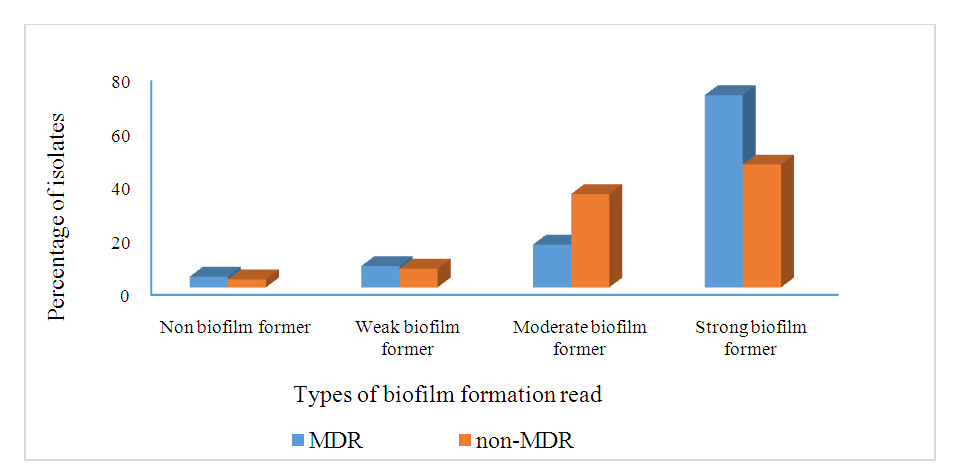 | Figure 4. The Relationship Between Biofilm Formation and Multidrug-Resistant isolates |
|
4. Discussions
- The primary focus of this study was to determine the total population and distribution of the Acinetobacter baumannii in leafy vegetables and clinical sources from Gondar, Ethiopia, including their antibiotic susceptibility, resistance genes, biofilm formation, and other associated virulence factors. Regarding the bacterial load of the vegetables, the mean aerobic mesophilic count of cauliflower, lettuce spinach and cabbage were 6.98cfu/g, 6.95cfu/g, 5.95cfu/g, and 4.95 cfu/g, respectively. These numbers are higher than previous reports by Akoachere in Cameroon, the mean was AMC of 1.68MPN/g for cauliflower, 1.50MPN/g for cabbage and 1.53MPN/g for spinach [51]. The higher bacterial load in this study may be partly due to the use of contaminated irrigation water and organic fertilizers in the farms, coupled with the poor hygienic environment where open defecation is not uncommon. Together these factors contribute to the microbial contamination during preharvest, harvesting, and poor handling practices at the post-harvesting [1,50]. These poor sanitary conditions were clearly observed when the vegetables were been sourced from the various markets. The maximum aerobic plate count (9.8 cfu/g) was from the vegetables obtained at Arada market while the lowest aerobic plate count was in the vegetables from Pizza market.To evaluate any causal relationship between the clinically relevant bacterial population from the vegetables and Gondar community hospital, the A. baumannii isolates from both sources were compared on the bases of their phylogenetic, phenotypic characteristics, plasmid profiling, antibiotic susceptibility, resistance genes and key virulence factors. The 16SrRNA sequencing and the phylogenetic relationship of the randomly selected isolates indicated the clinical and vegetable isolates features a relationship as they grouped together. As studies suggested that antibiotic resistance genes in human bacterial pathogens originate from different bacterial genetic sources including plasmid [52]. The genes carried in plasmids provide bacteria with genetic advantages, like antibiotic resistance. During this study, the plasmid profile for all selected isolates confirmed that every isolate had a minimum of one plasmid. Our result consistent with previous report from south India, the plasmid analysis revealed the presence of single or multiple plasmids in the nosocomial bacterial isolates as a carrier agent [53]. Besides their act as a carrier, these plasmids play a big role as a transferring agent to antibiotic resistant mobile genes [54].Regarding the antibiotic susceptibility the Acinetobacter baumannii isolates had high levels of antibiotic resistance (>98%). Previous findings were reported in northwest Ethiopia A. baumannii were resistant to Cefazolin (72.7%), Tetracycline (93.9%), and Ampicillin (67%). The overall prevalence of multidrug resistance (MDR) was 84% [6,84] which is lower than our report. Antibiotic resistant A. baumannii strains are prevalent and represent one of the most significant health-related problems in many areas of the world [3,4,5,30,47]. Studies conducted in Saudi Arabia and Greece in 2014 indicated extremely high percentages of antibiotic-resistant bacteria (>89%) to quinolone, beta-lactam/beta-lactamase inhibitor, cephalosporin, and carbapenem antibiotics [7,8,9,15]. Several studies have also indicated that colistin is the only remaining therapeutic option for multiple antibiotic-resistant Acinetobacter baumannii infections, with the percentage of resistant strains at less than 7.9%. However, A. baumannii is beginning to develop resistance to colistin in health care settings worldwide [13,14,32].Also, A. baumannii has become resistant to several classes of antibiotics due to the irrational use of antibiotics, leading to the predominance of multiple antibiotic resistant strains particularly in hospital settings [26]. Moreover, carbapenem resistance among A. baumannii isolates restricts therapeutic options for treatment of such infections which might lead to higher morbidity and mortality rates [41,43]. Comparing the results of resistant pattern for both clinical isolates which were tested for carbapenem antibiotics, (95%) were resistant to Doripenem (DOR), (89%) Imipenem (IPM), (89%) Meropenem (MEM), and (82%) Ertapenem (ETP). At present, our data is higher than previous reports among various resources in other countries [20,21]. On the other hand, for vegetable isolates that are tested for carbapenem antibiotics (84%) were resistant to Doripenem (DOR), (89%) Imipenem (IPM), (89%) Meropenem (MEM), and (82%) Ertapenem (ETP). For both clinical and vegetable isolates tested for 25 antibiotics, 99% were resistant to more than three classes of antibiotics. Most of the isolates were multiple antibiotics resistant to more than three classes of antibiotics. Also, isolates from vegetable sources and clinical samples were resistant to all common prescribed antibiotics in Gondar Ethiopia [22,25].On the other hand, genes encoding for blaOXA-51 and blaOXA-23 belonging to class D carbapenems were detected in 100% of the isolates, whereas ompA was detected in 30.7% of the isolates and ISAbaI is also found in 61.5% of the isolates. Numerous studies have lately reported that blaOXA-23 is the most prevalent carbapenems gene identified among A. baumannii isolates [31,33,85]. The prevalence rate of 100% reported in the current study agrees with previous studies. This study characterizing 27 A. baumannii isolates from Gondar Ethiopia showed a blaOXA-51, blaOXA-23 prevalence rate of 100 and 94%, respectively which is higher. The clinical isolates and vegetable isolates in both isolates are 98% resistant to imipenem and meropenem, respectively [27,28,29,32].Colonizing and biofilm formation ability of A. baumannii on biotic and abiotic surfaces is considered an important virulent factor [44]. Based on our results, all A. baumannii isolates were able to produce biofilm and 75% of isolates showed a strong ability to biofilm formation. Our results are consistent with previous reports which showed that more than 75% of A. baumannii isolates from biofilms [48]. Previous studies have reported a positive relationship between biofilm formation and antibiotic resistance during A. baumannii isolates [10,40]. In our study, all strong biofilm-forming A. baumannii isolates were MDR. Our study confirmed a significant correlation between strong biofilm formation and MDR phenotype (P < 0.01). Lots of reports from past studies have demonstrated that biofilm-related genes of A. baumannii including csuE, ompA, bap, epsA, bfmS were liable for biofilm development and antibiotic resistance [23,49].Regarding the analysis of virulence gene regions csgA, cnf1, cvaC, and iutA virulence genes had a substantial prevalence among the A. baumannii isolates of our samples. As previously reported by Daryanavard and Safaei [86] that the occurrence of csga, cnf1, cvaC, and iutA virulence genes among the samples of UTIs were 55%, 40%, 10%, and 30%, respectively which was like our findings. However, the past report by [36] which was on the occurrence of csga, cnf1, cvaC, and iutA virulence genes among the A. baumannii strains of clinical infections in Iran were 12.39%, 35.53%, 21.48%, and 19%, respectively was less than our results [35]. Al-Hassan showed that 40 isolates of A. baumanni strains of clinical infections had traT (80%), 17 isolates had cvaC (34%) and eight isolates had iutA (16%) genes [9]. As a virulence factor, these genes are the foremost common causes of adhesion and invasion of A. baumanni pathogens to the epithelial cells of the human organ systems [86].
5. Conclusions
- In the current study the aerobic mesophilic count and total coliform count indicated that the A. baumannii counts of all the vegetables exceeded the acceptable values set by the national standards of the country in assessing microbiological quality of certain fresh vegetables (>10, CFS, 2014; zero fecal coliform count FDA, 2006). All the isolates demonstrated a high prevalence of multiple antibiotic resistance A. baumannii in north Gondar Ethiopia. Carbapenem resistance genes were widely disseminated among Acinetobacter isolates. Carbapenem is that the last line of defense to treat infection with antibiotic resistant gram-negative bacteria, but the emergence of the carbapenem resistant genes has brought greater challenges to clinical treatment. In this study we identified the blaNDM-1, blaOXA23, blaOXA 51 and blaVIM resistance genes and csga, cnf1, cvaC, and iutA virulence factors for the first time in north Gondar Ethiopia. The results also revealed a genetic diversity among vegetable sample isolates of A. baumannii and clinical isolates from Gondar university hospital. The propagation of multiple antibiotic resistant species of A. baumannii among the isolates in both sources might be associated to the dissemination of the antibiotic resistance genes resulting in conventional failure of common antibiotic therapy treatments. Taken together, our findings demonstrated the presence of several plasmids associated to A. baumannii isolates which may contribute to the establishment of antibiotic resistance genes.
ACKNOWLEDGEMENTS
- Gratitude must be expressed to Dr. Rachelle Allen and Dr. Adrian Allen who has been helping me throughout this study, to Dr. Gugssa Ayele who assisted us in providing the resources for this project and Dr. Abdullah M. who assisted with technical support. Gondar University Department of Microbiology and Howard University Biology department, Applied microbiology laboratory.
Conflict of Interest
- There is no conflict of interest within any party in this research work.
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML