-
Paper Information
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
Journal of Microbiology Research
p-ISSN: 2166-5885 e-ISSN: 2166-5931
2019; 9(1): 12-24
doi:10.5923/j.microbiology.20190901.03

Potential Use of Soil Bacteria Associated with Potato Rhizosphere as Bio-control Agents for Effective Management of Bacterial Wilt Disease
Rostand Romeo Chamedjeu1, Joel Masanga2, Viviene Matiru1, 3, Steven Runo1, 2
1Department of Molecular Biology and Biotechnology, Pan-African University, Institute of Basic Sciences, Technology and Innovation
2Department of Biochemistry, Microbiology and Biotechnology, Kenyatta University, Nairobi, Kenya
3Department of Botany and Horticulture, Jomo Kenyatta University of Agriculture and Technology, Nairobi, Kenya
Correspondence to: Rostand Romeo Chamedjeu, Department of Molecular Biology and Biotechnology, Pan-African University, Institute of Basic Sciences, Technology and Innovation.
| Email: |  |
Copyright © 2019 The Author(s). Published by Scientific & Academic Publishing.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/

Potato has a crucial role to play in maintaining food security worldwide as it has high nutritive qualities and ratio of yield productivity to soil occupation. Despite the importance of the plant, its production is constrained by several biotic and abiotic stresses. Key among them is the bacterial wilt disease caused by Ralstonia solanacearum, with enormous yield losses. Existing management strategies have not been effective owing to the diversity of the pathogen with vast variation in host range. This study aimed at bio-prospecting for the potential of rhizopheral bacteria as biocontrol agents against potato bacterial wilt disease. The pathogen was isolated from plants showing typical symptoms of the disease and the obtained isolates were subjected to biochemical and molecular techniques of identification to confirm their status as Ralstonia solanacearum. In the course of bioprospecting, a total of 62 bacteria isolates were obtained from potato rhizosphere by serial dilution methods using broad spectrum media: nutrient agar and tryptic soy agar. Then, 20 isolates were selected based on their association with healthy plants for antagonistic tests using dual culture assay. During the in vitro screening, 5 bacteria species were identified to be highly antagonistic against four R. solanacearum strains. These antagonists were further tested in vivo for plant growth promoting traits and disease suppression ability. The results revealed that Bacillus cereus, Bacillus subtilis, Paenibacillus spp, Providencia rettgeri and Providencia vermicola were dominantly active in potato rhizosphere causing resistance to bacterial wilt disease. The isolates Bacillus cereus and Bacillus subtilis were also observed to have several plant growth promoting traits. These findings could provide baseline information for development of biocontrol strategies with the potential antagonists reported.
Keywords: Potato, Bacterial wilt, Ralstonia solanacearum, Biological control agents, Disease suppression, Growth promotion
Cite this paper: Rostand Romeo Chamedjeu, Joel Masanga, Viviene Matiru, Steven Runo, Potential Use of Soil Bacteria Associated with Potato Rhizosphere as Bio-control Agents for Effective Management of Bacterial Wilt Disease, Journal of Microbiology Research, Vol. 9 No. 1, 2019, pp. 12-24. doi: 10.5923/j.microbiology.20190901.03.
Article Outline
1. Introduction
- The whole world is faced with two diverse but interlinked challenges in the 21st century: feeding a growing population and how to adapt to climate change. During the last two decades, the number of undernourished people on the African continent has increased, indicating an increased importance of ensuring food security [1]. To meet the rising demand for food, potato cultivation has a role as a source of food with good balance of essential amino acids, protein and it produces an important quantity of energy per unit land than any other single crop (Abong et al., 2009; Ahuja et al., 2013). In addition, the Food and Agriculture Organization (FAO) proposed highlighting the potato for several reasons including its key role in the world global food system as it is the world’s fourth most produced food commodity, its ability to grow worldwide, its convenience for farming systems in developing countries with high ratio of yield productivity to soil occupation (85% of the plant is consumable compared with only 50% in cereals), and its nutritive qualities, with a higher amount of vitamins compared to grass plants [4]. These features make potato production a viable option that can significantly contribute to food security in Africa. Unfortunately, potato is susceptible to numerous abiotic and biotic threats such as bacterial wilt disease for which effective management techniques are yet to be identified. Therefore, the improvement of potato production relies in development of control strategies for the numerous microbial diseases that affect its growth, particularly bacterial wilt disease which remains an economically significant problem for smallholder farmers with losses estimated at about 50-75% [5]. This disease is ranked as the most devastating bacteria of potato and the Solanaceae family as whole (Kaguongo et al., 2010). It is the second most important potato disease after late blight [5], and has been estimated to affect about 1.7 million hectares of potatoes in around 80 countries [7], causing a global loss of more than USD 950 million per annum [8,9].Several management practices have been proposed and implemented for management of bacterial wilt disease. However, these have not been 100% effective mainly due the high diversity of the pathogen, its wide spread and host range [10]. Presently, to ensure food security, most agricultural systems are dependent on the use of chemical fertilizers and pesticides which are harmful to the environment, contribute to the destruction of soil microorganism, soil structure and diminishing the food quality [11]. Given these limitations, beneficial agricultural microorganisms used as biological control agents (BCAs) will be an important focus in pursuing sustainable agriculture while preserving the environment [12]. Several, studies on the potato rhizosphere, mycorrhizosphere and endorhiza have revealed the presence of a diverse and dense microbial community [9]. This microbial community constitutes a rich source for plant growth-promoting rhizobacteria and biocontrol agents. So far, the beneficial effects are related to microbial siderophores, antibiotics, biosynthesis of surfactants and phytohormones, nutrient and spatial competition, mycoparasitism, induced systemic resistance, phage therapy, quorum quenching and construction of transgenic lines [13].Several microorganisms have been described as possible biocontrol agents such as Hypericum gramineum, Pseudomonas flurescens and some species of Streptomyces [14]. Among the bacterial antagonists, many belong to the genus Bacillus and there are some other important genera such as Paenibacillus, Providencia and Trichoderma but are of lesser applied importance than Bacillus [15]. What make Bacillus species special is their unique ability to replicate rapidly, they are resistant to adverse environmental conditions as well as they have broad spectrum of biocontrol ability [16,17]. To a lesser extent, genera such as Arthrobacter, Comamonas, Curtobacterium, Enterobacter, Paenibacillus, Pantoea, Serratia, Sphingobacterium, Stenotrophomonas, Variovorax and Xanthomonas are also frequently found in the vicinity of the potato. Some of these species play an important role in plant growth promotion as they can synthetize plant growth-promoting hormones (GA3 and IAA) and defence-related enzymes (peroxidase (PO), polyphenol oxidase (PPO) and superoxide dismutase) [18].Biological control is among the most economical and eco-friendly disease management strategies. It provides an alternative safe method for control of disease and pests. It also represents an alternative to chemicals which have been reported to be hazardous to the environment as previously mentioned. One advantage of biological control using plant growth promoting bacteria (PGPB) is the reduction in the use of chemical agents against pathogens [9]. This minimizes problems associated with environmental pollution, ecosystem disruption and residual chemicals on crops as well as bioaccumulation of chemicals in the food chain. Therefore, knowledge of microbial community around healthy potato plants in an infested field is of special interest in development of biocontrol methods, which can limit recurrent losses and promote plant growth. This study was set up to screen in vitro for potential antagonists against R. solanacearum and to evaluate their disease suppression effects and growth promotion ability on potato plants.
2. Materials and Methods
2.1. Samples Collection, Isolation and Identification of Microorganisms
- Samples consisting of tubers, leaves, stems and soil around potato plants were collected randomly in three Kenyan sub-counties namely: Kuresoi North, Njoro and Mau Narok in Nakuru County where potato is mostly produced and the bacterial wilt disease is reported to be severed. Five samples each (infected plants, infected soil and soil underneath of healthy plant) were collected from each surveyed field for a total of five fields per sub-county. The pathogens were isolated from infected plants using tetrazolium chloride (TZC) medium as previously described by Kago and coworkers [19]. Virulent strains were selected based on characteristics of their colonies on TZC medium. These were confirmed to be R. solanacearum by DNA extraction using a Qiagen DNA extraction kit and amplification of common region of R. solanacearum genome using 759/760 primers [20].Other bacteria (potential antagonists) were isolated from soil collected underneath of healthy and infected potato plants, using serial dilution method. One gram of each soil sample was suspended in 9 ml sterile distilled water and diluted up to 10-4. Aliquots (100μl) of each suspension were spread on nutrient agar (NA) plates in triplicates and incubated at 28°C in an incubator. After 2 days of incubation, individual isolated bacteria colony was sub-cultured onto fresh NA plates. A total of 62 bacteria were isolated and purified. To avoid the harmful effects of the use of biological control agents (BCAs) to the plant, a screening assay for bacteria present only around healthy plant was performed on the core collection. This was done based on the color, shape and texture their colonies. Bacteria from non-infected and infected soil were cross-checked and 20 bacteria were selected for their presence in soil collected underneath of healthy plant and absence in an infected soil. These selected bacteria were then used for antagonistic activity screening against four R. solanacearum strains. Each isolate was preserved in 25% glycerol stock at -20°C for further experiments.
2.2. Inoculum Preparation for Antagonistic Assay
- Pre-cultures of bacteria were prepared by growing them in 10 ml of liquid nutrient broth (NB) for 24 hours at 28°C with agitation (200 rpm). R. solanacearum cells were cultured in Casamino Acid-Peptone-Glucose-agar (CPG) medium containing 0.1% Casamino Acids, 1% peptone, and 0.5% glucose at 28°C with shaking at 200 rpm. After growth, the medium was centrifuged at 5,000 g for 10 min at room temperature and bacterial cells were re-suspended in distilled sterile water. The optical density of samples from each tube was measured at 600 nm using a spectrophotometer (JENWAY 6300, Dunmow, UK) and the optical density (OD) was adjusted to 0.1 by adding more bacteria cells if the suspension was too light or diluting with sterile distilled water if the suspension was too heavy. Aliquots (100μl) of the dilutions were spread on nutrient agar plates and the colonies were counted after 24 hours of incubation at 28°C to estimate the density or growth of the bacterial cells.
2.3. In vitro Interactions of R. solanacearum with the Selected Bacteria
- The antagonistic activity of the selected bacteria against four R. solanacearum strains was assayed by dual culture (disc diffusion method) as described by Balouiri and his collaborators [21]. Paper discs of 6 mm diameter prepared from filter paper were autoclaved and impregnated with bacteria by soaking them in bacteria suspension and drying for 2-3 minutes. Discs impregnated with sterile distilled water were used as negative control. While the positive control entailed discs impregnated with gentamycin (10μg) and imipenem (10μg). Aliquots (100ul) of R. solanacearum suspensions (approximately 108 CFU/ml) was streaked on tryptic soy agar (TSA) plates a broad spectrum medium and 9 discs of each bacteria isolates were placed on top of the plates in triplicates. Plates were placed for 48 hours in the incubator set at 28°C. The interaction between R. solanacearum and the test isolates were monitored and a zone of inhibition around the paper disc was measured at day 2, 3 and 4 post-incubations. The experiment was repeated three times for accuracy.
2.4. Identification of the Bacteria with Significant Antagonistic Activity
- The isolates showing highly significant in vitro antagonistic activity were subjected to identification test using Api 20E kit following the manufacturer’s instructions. Bacterial identification was further performed through 16S rRNA gene sequences analysis. For this, bacterial cells grown for 1 day on nutrient agar (NA) were sub-cultured in Nutrient broth (peptone 5 g, sodium chloride 5 g, beef extract 1.5g, yeast extract 1.5 g, in 1L distilled water) and placed in a 28°C shaking incubator for 24 hours. Genomic DNA of all isolates was extracted using DNA extraction Kit (Qiagen) according to the manufacturer’s instructions. PCR reactions were performed using universal primers for bacteria: 27F (AGAGTTTGATCCTGGCTCAG) and 1492F (GGTTACCTTGTTACGACTT); and OneTaq 2X Master Mix with standard buffer (New England BioLabs) following the manufacturer’s recommendations. Thermal cycling parameters were as follows: a denaturation step at 94°C for 5 min followed by 35 cycles at 94°C for 30 s, 55°C for 30 s and 72°C for 90 s, with a final elongation step at 72°C for 7 min. PCR amplicons were verified by gel electrophoresis, purified using the GenElute PCR cleanup kit (Qiagen) and sequenced in both orientations at Inqaba biotech (Inqaba, SA). For each bacterial isolate, nucleotide sequences were trimmed, aligned and compared with the BLASTn search available in GenBank database. Phylogenetic relationships based on partial 16S rRNA gene sequences were determined with MEGA 7.0 software using maximum likelihood (ML) method with the General Time-Reversible plus gamma model of nucleotide substitution and bootstrap values of 1,000 interactions [22].
2.5. Evaluation of the Effects of BCAs on Plant Responses under R. solanacearum Infection
- The effect of the identified antagonists in inhibiting the growth of R. solanacearum cells in vitro led to evaluating their effects on plant response under R. solanacearum infection in planta. To do that, Certified tubers of S. tuberosum, “Shangi” variety from Potato Research Center (KALRO Tigoni, Kenya) were used for greenhouse experiments. This variety was chosen according to farmer’s preferences and it is known to be susceptible to bacterial wilt disease. The tubers were surface-sterilized with 0.5% sodium hypochlorite (NaOCl) and grown in pots containing double-sterilized combination of 4:1 ratio of soil and sand. The assays were conducted with five potential antagonists showing significant in vitro antagonistic activities against R. solanacearum. To prepare the inoculums, bacteria were grown on nutrient agar (NA) plates and a single colony was sub-culture in 10 ml of liquid nutrient broth (NB) for 48 hours at 28°C with agitation (200 rpm). The most virulent R. solanacearum strain (Rs6) selected for this experiment was cultured in CPG medium containing 0.1% Casamino Acids, 1% peptone, and 0.5% glucose at 28°C with shaking at 200 rpm. After growth, bacterial cultures with the medium (NB) were adjusted to an OD600 of 1 using a spectrophotometer (JENWAY 6300, Dunmow, UK). The OD was adjusted by adding more culture if the suspension was too light or diluting with sterile medium if the suspension was too heavy.Pot experiments in a greenhouse at plant transformation laboratory (PTL-Kenyatta University), were carried out following a complete randomized block design with 10 plants per treatment in triplicate. The experimental treatments were as follows: (1) control 1, containing plants with no biocontrol agent, no pathogen; (2) control 2, pots with no biocontrol agent but with infected soil; (3) A2 inoculation, in which the plants were treated with A2 isolate (108 cfu/ml); (4) A3 inoculation, in which the plants were treated with A3 isolate (108 cfu/ml); (5) A4A inoculation, in which the plants were treated with A4A isolate (108 cfu/ml); (6) A5 inoculation, in which the plants were treated with A5 isolate (108 cfu/ml); (7) A15 inoculation, in which the plants were treated with A15 isolate (108 cfu/ml); (8) co-inoculation1, in which the plants were treated with A2 and A3 combined (two bacteria which had shown inhibitory effect in vitro); (9) co-inoculation2, in which the plants were treated with A5 and A15 combined (two bacteria which had shown competition for space and nutrient in vitro) and (10) co-inoculation3, in which the plants were treated with A2 and A15 combined (two bacteria which had shown different antagonistic activities in vitro).All pots except that of control 1 were inoculated 24 hours before treatment, with 50 ml suspension of pathogen (Rs6, 108 cfu/ml). Then, the biocontrol agents were applied via root irrigation (seed treatment), a method developed by [23]. Ten pots were used for each treatment (for a total 100 pots per experiment), with each pot exposed to 50 ml of bacteria culture prepared as previously described. Plants were observed and wilt incidence was recorded weekly after the first wilting symptoms appeared. The stems of plants that did not show exhibiting wilt symptoms were also analyzed, to detect latent infections by plating their fragment on Modified Kelman’s Media [24].The treated plants were monitored for disease development for 30 days and disease incidence (DI) was calculated as DI (%) = 100 × (number of disease plants/10 inoculated plants) [25]. Biocontrol efficacy was calculated as [(disease incidence of control − disease incidence of treated plants) / disease incidence of control] ×100% [26]. After treatment, the effect of potential antagonists on plant growth was assessed in terms of germination percentage, plant height, fresh and dry biomass weight and water content. The growth promotion efficacy (GPE) was calculated to show the relative effect of antagonistic isolates on plant growth in comparison with that of control treatments by the following formula: Growth promotion efficacy (%) = [(Growth parameter in antagonist-treated group -Growth parameter in control group) / (Growth parameter in control group)] x 100 as reported by Almoneafy [27].
2.6. Data Analysis
- During the dual culture assays, the zone of inhibition was measured to define the interaction between tested bacteria and R. solanacearum strains. In greenhouse experiments, the identified potential antagonists were tested for plant growth promotion where germination percentage, plant height, fresh and dry biomass were measured. Disease suppression by the biocontrol agents were also evaluated by measuring the disease incidence and water content. These data were subjected to ANOVA analysis using R software and when the ANOVA was significant (P<0.05), the Tukey HSD multiple-comparison test was used for means comparison, with confidence interval specified through p-value. The graphical presentation of data was done using Graphpad prism 6 software.
3. Results
3.1. Isolated Microorganisms
- The pathogen was successfully isolated from diseased plant as the Kelman Tetrazolium Chloride (TZC) agar differentiation test gave colonies with pink or light red center and whitish margin for virulent isolates while avirulent isolates produced smaller, off-white and non-fluidal or dry colonies on TZC medium after 48 hours of incubation (Figure 1). Four isolates (Rs1A, Rs6, Rs15 and Rs35) displayed high virulence characteristics and were selected for further molecular characterization. The results from DNA-based analysis with 759/760 primers showed a PCR product of approximately 281bp. This confirmed that these isolates belong to R. solanacearum species.
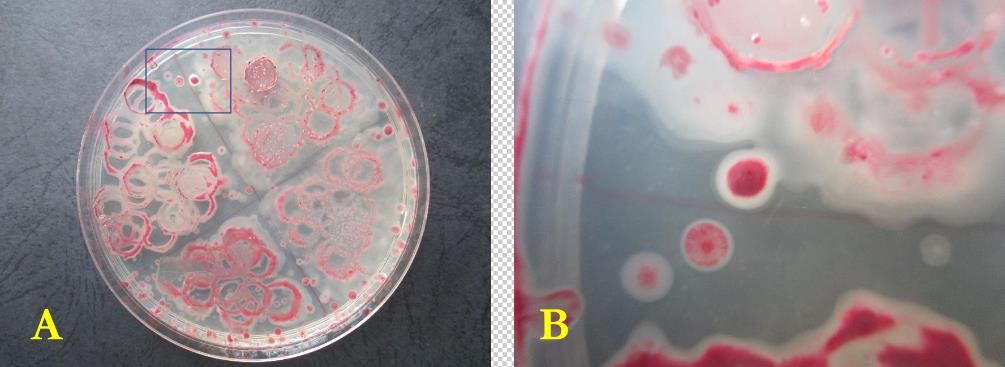 | Figure 1. A. Pure culture of R. solanacearum on TZC media, B. Zoom on virulent R. solanacearum colonies under microscope |
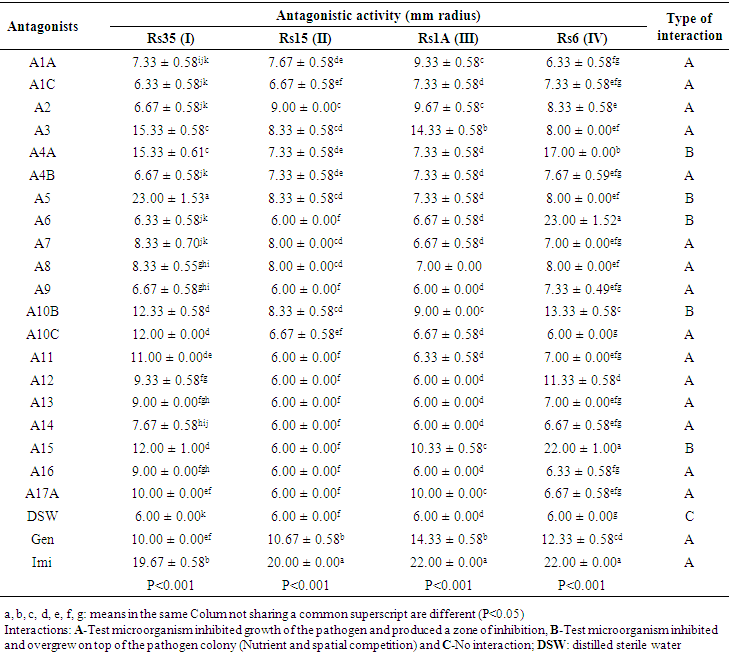 | Table 1. Antagonistic activity of the selected bacteria against four virulent strains of R. solanacearum |
3.2. In vitro Interactions of R. solanacearum with the Selected Bacteria
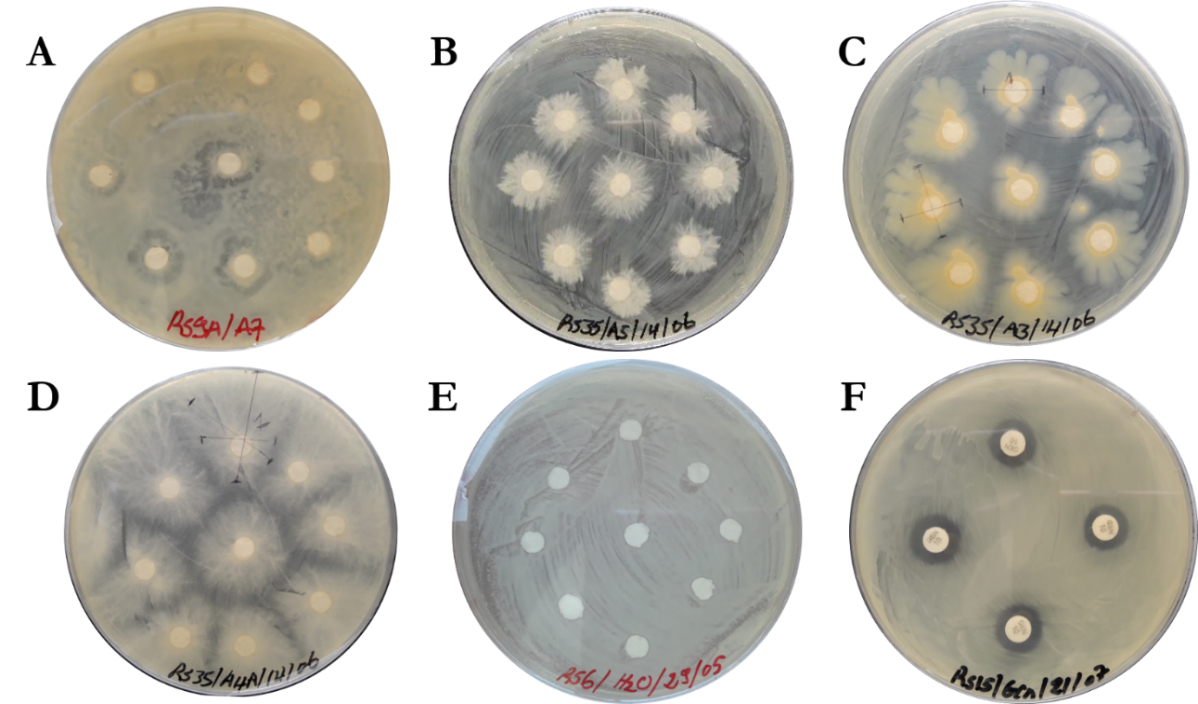
- Screening for rhizobacteria with antagonistic effects on four R. solanacearum strains in dual culture revealed 8 potential isolates (zone of inhibition > 8 mm in radius and space competition > 12 mm in radius) (Table 1). The tested bacteria had two types of interaction with the pathogen: most of antagonists inhibited growth of the pathogen and produced a zone of inhibition (antibiosis) while other inhibited and overgrew on top of the pathogen colonies (Nutrient and spatial competition) (Figure 2). In screening against Rs35, the highest antagonistic activity of 23.00 ± 1.53 mm was produced by A5 exhibiting competition for space and nutrient. The other isolates A3, A10C, A11, and A17A also showed significant inhibition results against Rs35 with highest inhibition zone of 15.33 ± 0.58 mm by A3 having antibiosis as mode of interaction. Similarly, in screening against Rs15, A2 was the best antagonist with the highest inhibition zone of 9.00 ± 0.00 mm followed by A3, A5 and A10B with inhibition zone of 8.33 ± 0.58 mm. Antagonism against Rs1A was recorded highest with A3 isolate having an inhibition zone of 14.33 ± 0.58 mm similar to that of gentamycin and isolate A15 represented moderate antagonistic activity of 10.33 ± 0.58 mm. Against isolate Rs6, A4A had the highest antagonizing potential with mean zone of inhibition of 17.00 ± 0.00 mm. Other isolates (A6 and A15) were also effective in competing for space and nutrient against Rs6. Interestingly, some antagonists showed greater inhibition zone compared to the positive control (Gentamycin) as shown on Table 1. Rs35 was the most susceptible among the pathogen strains tested since all antagonists inhibited its growth. On the other hand, Rs15 showed highest tolerance to the antagonistic effects of the tested microorganisms and only 13 antagonists were able to inhibit its growth.
3.3. Identification of Bacteria with Significant Antagonistic Activity
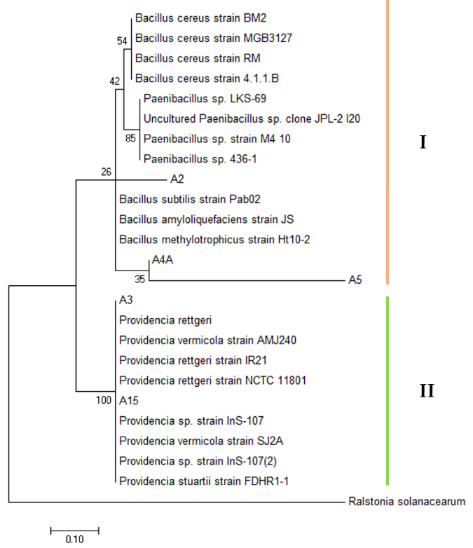
- Bacteria with significant in vitro activity were identified using 16S DNA sequences analysis. The basic local alignment search tool (BLAST) and phylogenetic analysis based on 16S rRNA gene sequences revealed that the potential antagonist with significant antagonistic activity belong to Bacillus cereus (A4A), Bacillus subtilis (A2), Paenibacillus sp (A5), Providencia rettgeri (A3) and Providencia vermicola (A15). These species were selected based on the lowest E-value, the highest query cover and identity percentage. Phylogenetic analysis grouped the bacteria into two cluster where Bacillus cereus, Bacillus subtilis and Paenibacillus sp. clustered together, while Providencia rettgeri and Providencia vermicola were grouped in the other cluster (Figure 3).
3.4. Evaluation of the Effects of BCAs on Plant Responses under R. solanacearum Infection
- From the in vitro assays, 5 potential antagonists were selected for green house experiment based on the type of interaction displayed against all tested R. solanacearum strains. The in vivo assays conducted under controlled greenhouse conditions were designed and tested beforehand to ensure a well-established and reliable pathosystem to allow the pathogen (Rs6) to grow and assure a complete infection of inoculated plants. The selected antagonists were evaluated for growth promotion (Figure 4 and Table 2), disease suppression (Figure 5) as well as for their efficacy (Figure 6 and Figure 7).
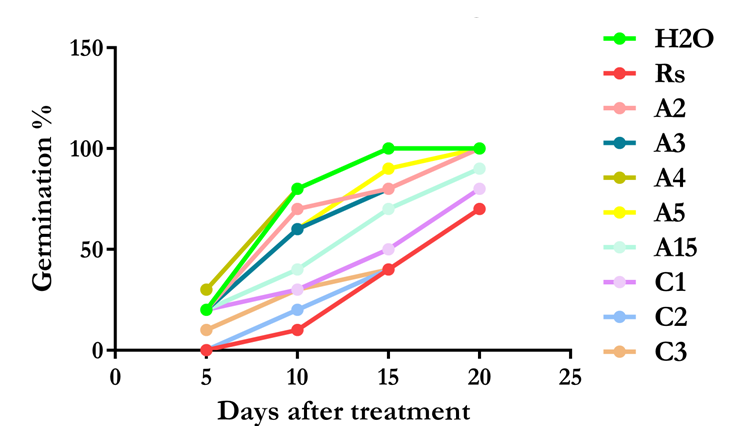
3.4.1. Growth Promotion Effects by the BCAs on Germination Percentage
- Plant were inoculated with the pathogen and the antagonists except in the negative control where only Rs6 was used and the positive control where water only was used. A growth promotion was observed on treated plants. The germination percentage recorded during the experiment demonstrated, that the treated plants emerged from the soil at early age as compared with the negative control (Without BCA application). All tested BCAs showed positive effects on the germination of the plants (Figure 4). Bacillus cereus (A4A) showed similar results as compared with the positive treatment (H2O), where the seeds were not treated but were sown in double sterile soil.
3.4.2. Growth Promotion Effects by the BCAs on Plant Height
- The growth promotion activity of the tested bacteria was also evaluated by measurement of plant height at four time points. The results from statistical analysis (ANOVA) performed for each tested biocontrol strain, confirmed the strong antagonistic activities of Bacillus cereus, Bacillus subtilis, Paenibacillus sp, Providencia rettgeri and Providencia vermicola against R. solanacearum on the sensitive potato variety “Shangi”. These bacteria significantly promoted the plant growth as compared to the negative control where only Rs6 were used (Rs). Interestingly, Bacillus cereus (A4A) promoted plant growth to the similar level than that of positive control with no infection (H2O) (Table 2).
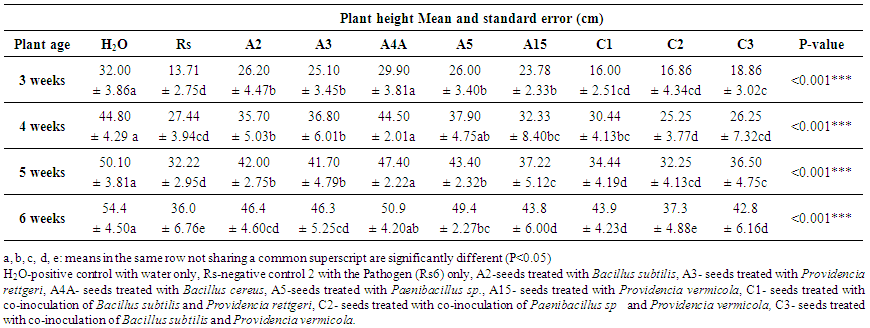 | Table 2. Effects of the identified BCAs on plant growth promotion (Plant height) |
3.4.3. Disease Suppression Activity of the BCAs
- In pot experiments, the tested BCAs showed some level of disease suppression as shown on Fig. 5. Bacterial wilt symptoms such as wilting appeared at 17 days post-treatments and were only observed on the negative control treatment with infected soil and not treated. A disease incidence of 20% was obtained in the negative control treatment. The Characteristics of bacterial wilt that were observed on diseased potato plants included: yellowing of leaves, stem streaks and wilting. None on these were observed on treated plants (Figure 6).
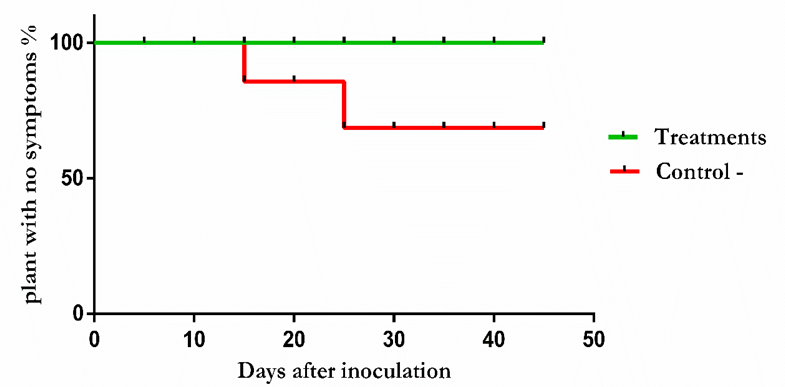 | Figure 5. Evaluation of the identified BCAs potentials in disease suppression |
3.4.4. Efficacy of the BCAs
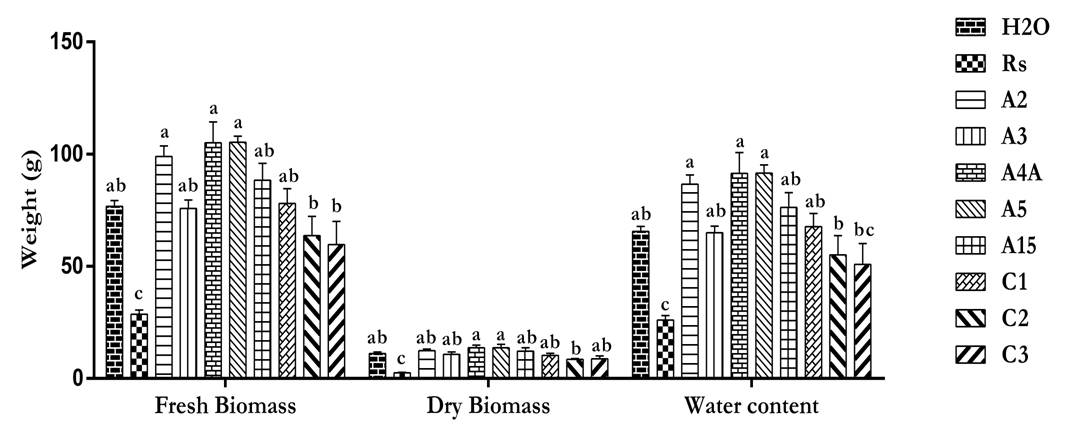
- Plant height was measured at four plant ages, the data showed that treated plants were taller compared to the non-treated. Bacillus cereus (A4A) had the highest effect on plant height compared to other individual and combined biocontrol agents (Table 2). Increase in the plant biomass were also observed with the treated plant as shown in Fig. 7. Water content was also used to evaluate the efficacy of the identified biocontrol agents, as bacterial wilt restricts the absorption of water and minerals. The result showed that, all treated plant had highly significant water content as compared to the negative control (Rs). This showed the ability of the BCAs to reduce infection and their properties to promote nutrient uptake, which are crucial for potato yield.
4. Discussion
4.1. Screening for Antagonists against R. solanacearum
- Potato rhizosphere have revealed the presence of a diverse and dense microbial community, where various candidates of antagonistic bacteria can be found with ability to affect the pathogen and promote plant development [28]. To select such potential antagonists, dual culture assay is required as a primary screening of bioactivity in vitro by providing information about interactions between a candidate antagonist and a pathogen on agar plate [29]. This is assessed by measuring the inhibitory and competitive effects of microbes on pathogen growth [30]. In this study, twenty (20) bacteria were selected and tested against four R. solanacearum strains. In general, five bacteria showed significant antagonistic activity against R. solanacearum. The results from this study demonstrated high significant variation among pathogen isolates in their susceptibility to the identified antagonists. This could be due to the high level of diversity both among the isolates of the pathogens and among the antagonists. A given antagonist showed different level of activity depending on the pathogen strains. The maximum inhibition was observed with Paenibacillus spp. when tested against Rs35, Bacillus subtilis against Rs15, Bacillus cereus against Rs6 and Providencia rettgeri when tested against Rs1A. This variation among pathogen isolates in their susceptibility to inhibition, was also observed by Otto-Hanson and his coworkers [31] when using antibiotic-producing isolates of Streptomyces sp. against Streptomyces scabies and the variation was dependent on the isolates of the plant pathogen.
4.2. Potential Mechanism used by the Antagonists to Inhibit the Pathogen’s Growth
- The bacteria used in this study had two types of activity against the pathogen in vitro; some had antibiosis activity where there synthesized certain molecules to inhibit the growth of the pathogen thus created an inhibition zone, while other competed for space and nutrient as they grown faster and over the pathogen colonies. These types of interaction have been reported in Bacillus genera as they are known to be appropriate candidates to be used in a bio-control approach due to their predominance in various environments, resilience and survival ability, but also for the number of bio-active molecules that they are potentially able to produce [32-34]. A study by Ashwini and Srividya [35] on B. subtilis isolated from chilli rhizosphere revealed appreciable levels of three mycolytic enzymes: chitinase, glucanase and cellulase which showed broad antagonism spectrum against potent bacterial and fungal phytopathogens. Additionally, many strains of B. subtilis have been reported for their chitinolytic activities [36].In this study, the suppressive activity of Paenibacillus spp. isolate was associated with antibiosis activity. This could be related to their ability to produce secondary metabolites that had effects on the growth of R. solanacearum. Paenibacillus spp. are also considered to be promising biocontrol agents of a number of plant diseases because of their wide host plant range, and ability to form endospores and produce various antibiotics [37]. In addition, antibiosis such as competition, root colonization and induced systemic resistance, have been associated with Paenibacillus spp and are proposed as the possible modes of action of these bacteria [38-40]. Similarly, the observed antagonistic characteristic of Providencia rettgeri against R. solanacearum, can be also related to its capacity to produce secondary metabolites such as acid from D-Adonitol, D-Arabitol, Erythritol, and other metabolic precursors, and reduction of nitrate to nitrite. P. rettgeri has been more effective in competition for space and nutrient, this can be related to its ability to convert amonia to nitrogen as reported by Zhao and his collaborators [41].
4.3. Efficiency of Biological Control Agents (BCAs) against R. solanacearum
- The dual culture assay provided an insight into the level and range of the tested bacteria bioactivity on R. solanacearum strains. Even though, in vitro antagonisms are not always a good indicator of biocontrol activity in vivo [42]. This is because, many parameters such as age, temperature, pH, and nutrient composition of the media can affect metabolite production by an organism and can result in different bioactivity outcomes in an in vitro study. These bioactivities were observed in vivo in term of growth promotion and disease suppression on plants grown under disease infection.
4.3.1. Growth Promotion Activity of the Tested BCAs
- In the pot experiment, some treatments indicated significant increase of plant growth parameters and reduction of disease incidence while others did not. The treatment of seeds with the identified antagonists, prevented the development of wilt symptoms and significantly promoted plant growth in relative to the non-treated plants. This is in line with Kamil’s findings [43], where they reported that sunflower seeds coated with B. licheniformis induced high reduction in percentage of infection of Rhizoctonia solani damping-off (from 60 to 25%) as compared to the pathogen alone. The increase of plant growth parameters by seed treatment with biocontrol agents is also an observation made in previous studies by Jung [44] and Khan [45]. The reason for the increase development may be attributed to synthesis of plant hormones such as cytokinin and auxin [46]; and facilitation of nutrient availability through nitrogen and phosphate metabolism [47]. Rhizobacteria (in particular PGPR) also known to act as biofertilizer can promote plant growth by breaking down soil complexes and suppression of the deleterious effects of abiotic and biotic stresses [48]. The application of chemical nitrogen and phosphorus fertilizer could be reduced by inoculating fields with phosphorus-solubilizing microorganisms, such as Paenibacillus, Bacillus and Providencia species [49,50,17].The ability of the identified antagonists including Paenibacillus, Bacillus species to produce auxin and gibberellic acid (GA) demonstrates the huge potential of these microbes as BCAs as well as plant growth enhancers [51,52]. Auxins and gibberellic acid (GA3) are hormones that are crucial regulators of gene expression and development throughout a plant’s life, participating in cell division, elongation, fruit development and senescence [53]. These plant growth-promoting hormones also enhance the nutrients uptake ability of plants and help the plant to defend against various biotic and abiotic stresses [54,55]. Apart from the antagonistic mechanism of Bacillus species, these microbes also have an important role in plant growth promotion by enhancing the biosynthesis of plant hormones gibberellic acid (GA3) and indole-3 acetic acid (IAA) that have a close relation with plant nutrient availability [39].
4.3.2. BCAs’ Ability in Suppression of Bacterial-wilt Disease Associated Symptoms
- In this study, the tested BCAs significantly reduced the disease incidence as compared to the non-treated plant. Disease suppression mechanisms have been reported by Wang [56], who concluded that treatment with B. cereus (strain AR156) enhances defense-related activities such as phenylalanine ammonia-lyase (PAL), chitinase, β-1,3-glucanase, peroxidase (PO), polyphenol oxidase (PPO), and stimulated amassing of Hydrogen peroxide (H2O2). By producing biocidal substances, they can induce the plant resistance mechanisms and neutralize a diverse variety of phytopathogens and insect herbivores [40]. Park and his coworkers [25] evaluated five strains of Bacillus species against R. solanacearum and the studied strains were proved to be effective. They also showed that reduction of disease was not due to direct antagonism but as a result of elicitation of host plant resistance genes. Similarly, Paenibacillus have been reported to control phytopathogens by triggering induced systemic resistance (ISR) and/or producing a variety of biocidal substances [40].Beneficial nonpathogenic microorganisms can induce systemic acquired resistance (SAR), a phenomenon where a non-infected plant acquires an ability to resist the subsequent attack. This is seen when colonization of roots by some nonpathogenic bacteria protects the above-ground plant parts from attack of various pathogenic organisms, which is known as induced systemic resistance (ISR) [57]. Bacillus, Paenibacillus, and providencia species are beneficial rhizobacteria and root-associated mutualists which can trigger ISR when present in high enough population densities [58]. Previous studies have investigated the elicitation abilities of B. subtilis, B. cereus and other Bacillus strains to induce a broad spectrum of resistance against various bacterial and fungal phytopathogens [32,59].The disease suppression effect by Providencia species, as shown in the result part can be explain by the high interspecific competition between these bacteria and the pathogen causing reduction in the growth, productiveness and other activities of the pathogen. Biological control can be observed when pathogenic and nonpathogenic organisms compete for space and nutrients around the host plant [60]. In general, soil-borne plant pathogens such as R. solanacearum are more exposed to competition because they only infect through infiltration of plant tissues in comparison with those pathogenic diseases whose causal organisms can directly germinate on plant surface. Therefore, non-pathogenic microbes can protect plants by rapid colonization and exhausting the developmental resources thus making them unavailable for pathogenic microbes [17].
5. Conclusions
- This study confirmed the positive effects of certain bacteria associated with potato rhizosphere, in suppressing bacterial wilt disease and boosting plant development. Five bacteria were identified to be highly antagonistic to R. solanacearum and should be taken in consideration in formulation of biocontrol agents for bacterial wilt disease. There is need to understand why these bacteria were working efficiently. Therefore, further study need to be done to identify the mechanism used by the bacteria to suppress the disease and there is also need to evaluate the BCAs under different field conditions before real recommendation in mass production of bio-fertilizers.
ACKNOWLEDGEMENTS
- The authors acknowledge financial support from the African Union through Pan African University. We are grateful to the Ministry of Agriculture of Kenya for facilitating sample collection. We also acknowledge the technical assistance provided by the International Potato Center (CIP).
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML