-
Paper Information
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
Journal of Microbiology Research
p-ISSN: 2166-5885 e-ISSN: 2166-5931
2018; 8(4): 97-102
doi:10.5923/j.microbiology.20180804.02

Molecular Characterization and Antimicrobial Efficacy of Streptomyces gancidicus Strain SN-3 from Soil Samples
Sreenivasa Nayaka1, Chethan J. Dandin2, Girish Babu K.1, Ravichandra Hospet1, Pallavi S. S.1, Bidhayak Chakaraborty1
1Department of P.G. Studies Botany, Karnatak University, Dharwad, Karnataka, India
2Department of P.G. Studies in Microbiology and Biotechnology, Karnatak University, Dharwad, Karnataka, India
Correspondence to: Sreenivasa Nayaka, Department of P.G. Studies Botany, Karnatak University, Dharwad, Karnataka, India.
| Email: |  |
Copyright © 2018 The Author(s). Published by Scientific & Academic Publishing.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/

The present work was carried out for four Actinomycetes SN-1, SN-2, SN-3 and SN-4 isolated from soil samples of agricultural fields near University of Agricultural Sciences, Dharwad, Karnataka. Isolates were subjected for the screening of antimicrobial analysis by the cross streak method. Among four isolates the strain SN-3 has shown good antimicrobial activity. Morphological characterization by scanning electron microscopic analysis followed by physiological and biochemical characterizations were performed. The potent strain was identified by the 16S rRNA gene sequence (987 bp) and a phylogenetic tree was correlated with other species Streptomyces spp and finally, the strain was confirmed and identified as Streptomyces gancidicus SN-3.
Keywords: Actinobacteria, Streptomyces gancidicus, Antimicrobial, 16S rRNA
Cite this paper: Sreenivasa Nayaka, Chethan J. Dandin, Girish Babu K., Ravichandra Hospet, Pallavi S. S., Bidhayak Chakaraborty, Molecular Characterization and Antimicrobial Efficacy of Streptomyces gancidicus Strain SN-3 from Soil Samples, Journal of Microbiology Research, Vol. 8 No. 4, 2018, pp. 97-102. doi: 10.5923/j.microbiology.20180804.02.
Article Outline
1. Introduction
- Soil considered as a good source of potent microorganisms and is an excellent resource for identification and characterization of novel antibiotic producing microorganisms among which Actinomycetes are known for their potential features in the production of various antibiotics. Actinomycetes are called Actinobacteria with high guanine and cytosine content present in DNA. The Actinobacteria are gram-positive bacteria and capable of producing antibiotics, secondary metabolites and bioactive compounds. In microbes approximately 45% of the bioactive compounds produced from the Actinomycetes [1]. In the world, 80% of antibiotics are available by of the genera Micromonospora and Streptomyces sp [2]. The Actinomycetes have been isolated and identified from many ecological origins such as soil, marine and freshwater bodies, compost, marshy places and extreme environments. Among these natural medium, soil witness the greatest Actinomycetes population density. The extreme environment such as acidic, alkaline, high temperature and pressure, complex ecosystems and the rhizospheres can be observed for screening of new Actinomycetes [3-5]. Actinobacteria are filamentous like structure, filaments are long and it may fragment into smaller units. Identification of Actinomycetes filamentous structure using microscopic and scanning electron microscope is not enough further, the biochemical methods help in identification of isolates for genus level. Presently with advanced technology, the many researchers performing 16S rRNA sequence analysis of the Actinomycetes isolates for the species level identification [6,7]. In the recent years noticing that the rare Actinomycetes are isolated from various soil samples from unexplored as well as uncommon extreme environments [8, 9]. In the present study, we have focused on isolation, morphological, physiological, biochemical and molecular characterization of Actinomycetes from soil samples.
2. Materials and Methods
2.1. Collection of Soil Samples
- Soil samples were collected at various locations from agriculture areas nearby Agriculture University, Dharwad, Karnataka. The selection of field study area was followed as per the standard protocol by Skinner [10]. Total of 13 soil test samples were collected and removed the stones and debris from it. Further, soil test samples were dried for four to five days at normal temperature and then refilled in respective polyethylene bags and kept at 4°C for the further studies. The isolation of Actinomycetes was performed by employing a standard protocol of serial dilution plate method [11]. Standard serial dilution of 10-1 to 10-5 was prepared from the test soil samples. The sample of 0.1 ml from respective dilutions was spread on the starch casein agar plates [12]. The plates were incubated for one week at 30°C. The growth activity of Actinomycetes was observed on plates for every 24h.
2.2. Morphological Characterization of Actinomycetes
- The morphological features such as colony shape, aerial hyphae, mycelium and spore formation of the selected culturing Actinomycetes isolates strain was examined on starch casein agar medium [13].
2.3. Physiological and Biochemical Characterization of Actinomycetes
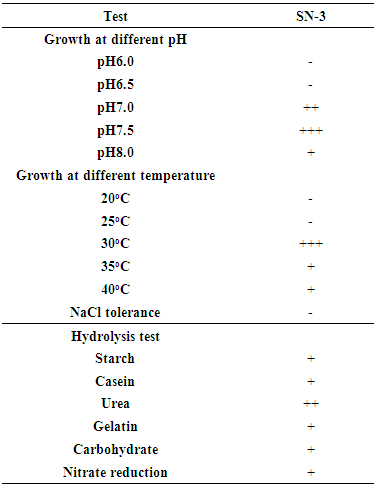
- Physiological and biochemical characterization of elite isolates were studied to understand the basic physiology of the soil Actinomycete. Several physiological and biochemical tests analysis such as growth at different pH and temperature, NaCl tolerance, gram and spore staining, motility test, starch, urea, casein, carbohydrate and nitrate reduction analysis were examined [14].
2.4. Primary Screening for Antimicrobial Activities of Pure Isolates by Cross Streak Method
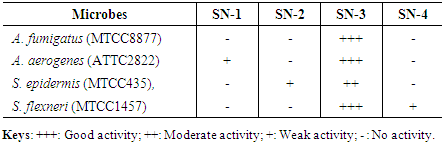
- Isolated strains were subjected for evaluation of antimicrobial potential. Pathogenic bacteria such as A. fumigatus (MTCC8877), A. aerogenes (ATTC2822), S. epidermis (MTCC435) and S. flexneri (MTCC1457) were procured from CSIR-IMTECH Chandigarh, India. Nutrient agar media was used for the antimicrobial activity and cross streak method was followed [15].
2.5. Scanning Electron Microscopy Analysis
- After the preliminary examination of antimicrobial activity, a unique isolate which exhibited potential antimicrobial activity was further subjected for morphology and external morphology studies using scanning electron microscopy and a sample was prepared according to the protocol described by [16].
2.6. DNA Extraction and Analysis of 16S rRNA and DNA Sequence
- Genomic DNA was isolated [17]. Briefly, 1.5 ml of 24h old isolate was centrifuged at 10,000 rpm for 3 min to get pellet and 0.5 ml of lysis buffer was added (100 mM Tris, 10% SDS, 50 mM EDTA, pH 8.0). Further, 0.2 ml chloroform and 0.5 ml of phenol was added and kept at 55oC for 10 min. After incubation, centrifuged for 10 min at 10,000 rpm and to the supernatant equal volume of chloroform: isoamyl alcohol (24:1) and 1/20th volume of 3 M sodium acetate was added. Later centrifuged for 10 min at 10,000 rpm and to supernatant 3 volumes of pre-chilled ethanol was added. Precipitated DNA was recovered by the centrifugation. Pellet containing DNA was air dried and dissolved in the TE buffer (10 mM Tris HCL, 1 mM EDTA, pH 8.0) and stored in freeze at 4°C. The quality of DNA was analyzed by the performing agarose gel electrophoresis and NanoDrop analysis. The 16S rRNA gene segments were amplified using primers GAAGCGCTCACGGCCTA (Forward Primer) and CGGAGTGTCCATGTTCAGGGAACG (reverse primer). The PCR reaction mixture contains 50 ng of DNA, 1 μl of 16S forward primer of 400ng and 16S reverse primer of 400ng, 4 μl of dNTPs (2.5 mM each), 10 μl of 10X Taq DNA polymerase buffer, 1 μl of Taq DNA polymerase (3U/μl), and millipore water was added to make the total reaction volume to 100 μl, amplification was performed using automated thermal cycler (Model; ABI2720, Applied Biosystems USA), with following conditions; initial denaturation was performed for 5 min (25 cycles) at 96°C, followed by denaturation at 96°C for 30 sec, Extension at 50°C for 30 sec and the final extension performed at 60°C for 1.30 min. The PCR amplicons were examined on agarose gel electrophoresis (1%) with the aid of 500 bp DNA ladder. The purified PCR product was sequenced (applied biosystems Sanger sequencing 3500 series genetic analyzer) and examined to interrogate at NCBI database using BLAST analysis [18].
2.7. 16S rRNA Phylogenetic Tree Analysis
- Sequenced DNA data were compiled and analyzed and matched with the GenBank database using a Basic local alignment search tool (BLAST) algorithm from the NCBI website (www.ncbi.nlm.nih.gov). The genomic DNA sequence was efficiently aligned, analyzed for the species identification and the phylogenetic tree was built by the neighbour joining method using the software MEGA4 [19]. The SN-3 culture was deposited in the gene bank and the accession number is KX284896.
3. Results
3.1. Identification and Morphological Characterization of Actinomycetes
- The pure and clearly distinguishable colonies of 4 Actinomycetes isolated from 13 soils samples, these strains are slow growing and shows different colony colours like grey, white, orange and brown. The scanning electron microscope of selected strain SN-3 examined for spore chain and it was a cubic form with 10 to 20 forms or more than that (Figure 1).
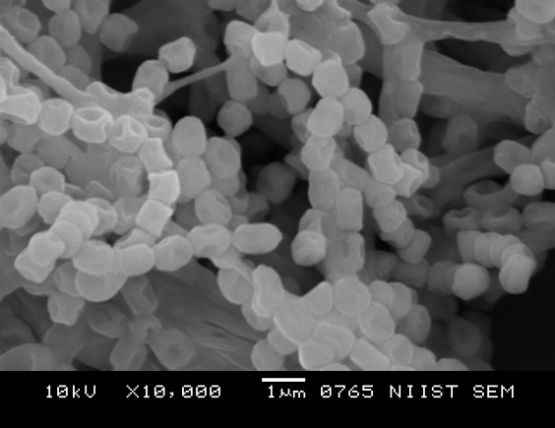 | Figure 1. Under Scanning Electron Microscope Examined Spore View for Selected Isolate SN-3 |
3.2. Antimicrobial Activity
- Four isolates used for the screening of antimicrobial activity on nutrient agar plates using cross streak plate method. The inhibition zones of the Actinomycete isolates against the different pathogenic microorganisms were observed as illustrated in the Table 1. The Actinomycetes culture SN-3, only one isolate showed a broad effect of antimicrobial activity of both gram-positive and gram-negative bacteria. On the basis of the broad effect of antimicrobial activity the SN-3 isolate used for furtherer work.
|
3.3. Physiological and Biochemical Characterization
- The physiological characterization of isolate SN-3 growth of pH adjusted at 5.0 to 8.0 and temperature was adjusted at 20°C to 40°C. The growth activity of isolate SN-3 was optimum at pH 7.5 and the temperature 28°C was observed for relative good growth. The biochemical characterization of isolate SN-3 included the utilization of starch, casein, urea, gelatine and nitrate reductions except for carbohydrate and NaCl is not utilized by SN-3 actinomycetes. (Table 2).
|
3.4. 16S rRNA Gene Sequencing
- 16S rRNA sequencing is one of the elegant methods for the identification of soil microbes; the gene sequencing of isolate SN-3 was of findings found 987 bp (Figure 2). The phylogenetic tree clearly indicates its evolutionary relationship with a group of Streptomyces sp. The BLAST analysis of the 16S rRNA sequencing of a strain of SN-3 was performed and compared with most similar sequence results of Streptomyces gancidicus strain 15412 and Streptomyces gancidicus strain AG. Consequently, the isolate was identified and confirmed as Streptomyces gancidicus SN-3 further, a sequence was deposited in the NCBI GenBank database and got the accession number as KX284896 (Figure 3).
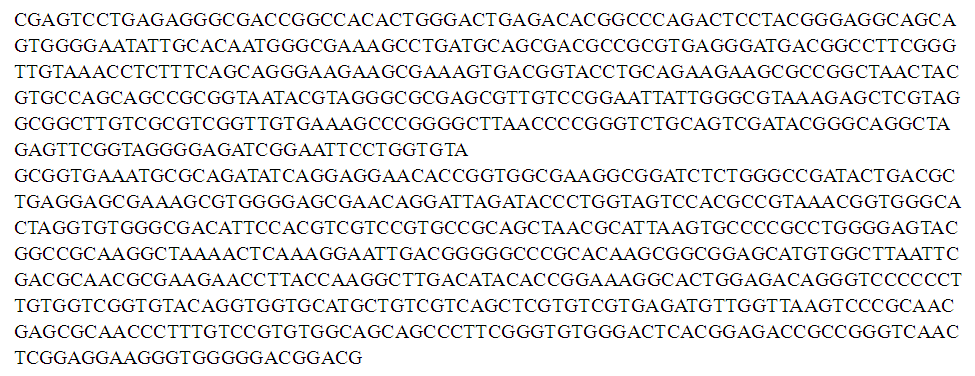 | Figure 2. Complete 16S rRNA Gene Sequence of the Streptomyces gancidicus SN-3 (987 bp) |
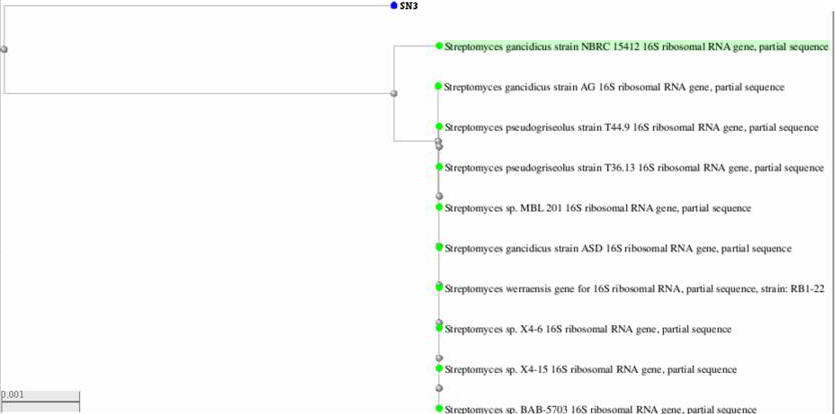 | Figure 3. Neighbor Joining Tree of 987 bp of 16S rRNA Gene Sequences of Streptomyces gancidicus SN-3 Isolate (GenBank accession no. KX284896) |
4. Discussion
- Actinomycetes are isolated from several unexplored soil regions from different parts of the world in recent studies. In the current studies Actinomycetes are isolated from the soil samples of agricultural fields and based on gram staining and colony morphology, totally four Actinomycetes are confirmed for further analysis. The similar observation was done in thirty-seven Actinomycetes strains isolated from soil samples from an agriculture field in Vengod [20]. In 15 soil samples thirty different Actinomycetes were observed in Gondar town [21]. Many similar observations were reported such as, 31 Actinomycetes strains were isolated from soil samples from sheopur [22], and Ninety-seven Actinomycetes strains were isolated from fifty soil samples collected from the Taif City, Saudi Arabia [23]. In the region of Egypt one hundred soil samples are collected and isolated 12 Actinomycetes [24]. Among four isolates SN1, SN2, SN3 and SN4 only one isolate SN3 shows the broad range for the initial screening of antimicrobial activity, further these isolates were subjected for the characterization based on the morphological, physiological and biochemical analysis. Similar results were reported earlier, 20 isolates out of 150 Actinomycetes [25, 26] and 32 out of the 320 Actinomycete strains from various Moroccan habitats shown strong activity against moulds, yeast and bacteria [27]. Jain and Jain [28] Out of 287 Actinomycetes 166 isolates were found antagonistic properties against different pathogenic organisms.In several reports it was observed that phylogenetic tree analysis based on sequencing of 16S rRNA gene has served as an important tool for identification of Actinomycetes. 16S rRNA sequence was determined and BLAST analysis was performed, the active strain SN-3 was 90% percent similar to Streptomyces gancidicusa NBRC1541, Streptomyces gancidicus AG and Streptomyces Gancidicus T449. Based on the all experimental data finally, the strain SN-3 was confirmed as the Streptomyces gancidicus SN-3. Similar results were reported earlier in 16S rRNA gene sequencing as a powerful method for identification of prokaryotic organisms [29]. The number of Actinomycetes are identified by sequencing the 16S rRNA gene as Streptomyces [30]. Streptomyces albidoflavus [31], Nocardioides [32] up to species level.
5. Conclusions
- The present investigation was carried out on isolation and functional screening of potential soil Actinomycetes for potent antimicrobial properties. Among four isolates Streptomyces gancidicus SN-3 shown optimum antimicrobial activity and it was subjected for various analysis for its identification. Such soil Streptomyces sp play a vital role in the production of various classes of secondary metabolites. Hence the isolated strain Streptomyces gancidicus SN-3 has to be subject for further analysis for its ability in quality production of antibiotics and biological control of plant diseases caused by many phytopathogenic fungi and also be screened for synthesis of nanoparticles for its potential applications in nanoagriculture as such application are greatly encouraged nowadays.
ACKNOWLEDGEMENTS
- The authors thank the Chairman of the Postgraduate Department of Studies in Botany, Karnatak University, for providing the necessary facilities and UGC–DSA–I: University Grant Commission Departmental Special Assistance–I phase program of the Department of Botany, Karnatak University, Dharwad Campus.
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML