-
Paper Information
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
Journal of Microbiology Research
p-ISSN: 2166-5885 e-ISSN: 2166-5931
2017; 7(4): 93-98
doi:10.5923/j.microbiology.20170704.02

Research of ESBL in Bacteria Responsible for Pediatric Urinary Tract Infection
Khaldi A. 1, Tifrit A. 1, Moussa K. 2, Benine A. 1, Abbouni B. 1, Tir Touil A. 2, Meddah B. 2, Benali M. 3
1Laboratoire de Microbiologie Moléculaire, Protéomics et Santéuniversité de Sidi Bel Abbes, Algérie
2Laboratoire de Recherche Sur le Système Biologique et Géomatique Faculté SNV, Université Mustapha Stambouli Mascara, Algérie
3Laboratoire de Biotoxicologie Faculté de SNV Université de Sidi Bel Abbes
Correspondence to: Khaldi A. , Laboratoire de Microbiologie Moléculaire, Protéomics et Santéuniversité de Sidi Bel Abbes, Algérie.
| Email: |  |
Copyright © 2017 Scientific & Academic Publishing. All Rights Reserved.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/

We studied pediatric antibiotic resistance and evaluated the impact of antibiotic exposure in the previous 10 months, with very little available Algerian data for this population. We conducted a prospective, multicenter study involving children who were admitted or admitted to the sidi bel abbes hospital in western Algeria. Pre-exposure to antibiotics was documented from their medical records. Of 75 patients (40 girls), 2 to 12 years, were included in 10 months. 75% percent had pyelonephritis associated with 25% uropathy. Escherichia coli was predominant (78%). In Gram-positive bacteria, Staphylococcus aureus was most frequently isolated with 23%. The antibiotic resistance rate of E. coli was high and close to that reported for adults with complications: 60% amoxicillin, 35% amoxicillin-clavulanate, 5% cefotaxim, 26% sulfametoxazole trimethoprim, 9% nalidixic acid, ciprofloxacin 7%, Gentamycin 1%, nitrofurantoin and fosfomycin 0%. Exposure to antibiotics in the past 12 months was most frequent with 62 children (56%) with β-lactams (89%) for respiratory tract infection (56%). The bacterial species and the level of resistance to antibiotics in children are similar to those reported in adults. Our results indicate that all strains isolated and identified in various sites were multiresistant. Antibiotics 50% of strains isolated from Staphylococcus aureus are presumed to be MRSA. In the Gram Negative and Enterobacteriaceae 79.46% isolated strains producing ESBL. Exposure to antibiotics in the past 12 months increases the risk of resistance but other factors are involved (previous antibiotic therapies and fecal-oral or mother-to-child transmission).
Keywords: Bacterial resistance, Pediatric Urinary, Tract infection
Cite this paper: Khaldi A. , Tifrit A. , Moussa K. , Benine A. , Abbouni B. , Tir Touil A. , Meddah B. , Benali M. , Research of ESBL in Bacteria Responsible for Pediatric Urinary Tract Infection, Journal of Microbiology Research, Vol. 7 No. 4, 2017, pp. 93-98. doi: 10.5923/j.microbiology.20170704.02.
1. Introduction
- For years, urinary tract infections in young people have been a real public health problem encountered in both city and hospital practices, partly due to antibiotic treatment and partly to their clinical diagnosis based on macroscopic examination. Urine strips (LAPSTIX / MULTISTIX), carried out in the doctor's office allowing immediate therapy, but this remains insufficient and is affirmed only by a cytobacteriological examination of urine (ECBU).The selection of bacteria developing antimicrobial resistance systems against these molecules has considerably modified the microbial ecology that is manifested by their ability to appropriate new properties either by modifying their genome (mutations) or by acquiring new genetic information. Thus, the dissemination of resistance genes between bacteria has led to the appearance of BMR (multi-resistant bacteria), in particular strains of MRSA (methicillin-resistant staphylococcus aureus) and EBLSE (Spectrum β-lactamase-producing enterobacteria). extended).This public health problem amply justifies efforts to understand this epidemiology.At the level of the wilaya of Sidi Bel Abbes, to our knowledge, no study has been done on urinary tract infections in hospital pediatrics which justifies the realization of this work carried out in the University Hospital - AEK HASSANI in order to specify the epidemiological data, clinical and therapeutic.
2. Materials and Methods
- Samples collectionThis study was conducted between March and December 2016 from the hospital of sidi bel abbes western north of Algeria. A total of 75 patients (40 girls), 2 to 12 years of age, 75% presented with pyelonephritis, associated to uropathy for 25%.The urines were transported to the laboratory and stored at – 20°C until further analysis.Bacterial isolation and identificationAll samples from urinary infections, were prepared by diluting scraped cell mass in 0.85% NaCl sterile solution and then analyzed for aerobic bacterial content by cultures on a series of non-selective and selective media (Blood agar, Chapman medium, Hektoen medium, Nutritive agar medium. Plates were incubated at 37°C for 24 and 48 h, grown colonies and pure cultures were identified by recommendation of Bergy’s [6] using standard morphological, Gram coloration and biochemical methods with commercials kits (API Staph, API 20 E Biomerieux, Marcy l’Etoile, France). The tests were conducted twice for each sample and the mean of Colony Forming Unit (CFU) count was determined. The microbial strains were maintained on agar slant at +4°C until analysis.Inoculums’ preparationNutriment broth [7] was used for growing strains and diluting suspensions. Bacterial strains were grown to exponential phase in nutriment broth at 37°C for 18 h and adjusted to a final density of 2x108 CFU by diluting fresh cultures and comparison to Mac Farland standards (OD650=0.7) [8].Antibiotic susceptibility testingResistance towards antibiotics was assessed for each strain with the disc diffusion method [9] and bacterial growth on Muller Hilton Agar plates. The antibiotic tested for Enterobacteriaceae were Ampicillin (10 μg), Gentamicin (10 μg), Aztreonam (30 μg), Colistin (10 μg), Tetracyclin (30 μg), Chloramphenicol (30 μg) Amoxicilline (), cefotaxin Acide nalidixique () ciprofloxacine amoxicilline – clavulanate () trimethoprime-sulfametoxazole () for Staphylococaceae, were Oxacillin (10 μg), Erythromycin (15 μg), Spriramycin (10 μg), Chloramphenicol (30 μg), Tetracyclin (30 μg), Staphylococcus aureus resistant Methicillin (MRSA) searchDip a sterile swab into the bacterial suspension standardized at 10⁶ CFU / ml. The swab is rubbed on the agar surface of the screen (Muller Hinton). Submit a oxacillin 10 μg the center of the box hard and incubated for 24 h at 37°C isolated in the presence of the inhibition zone around the disk colonies resistant MRSA and heterogeneous [10].
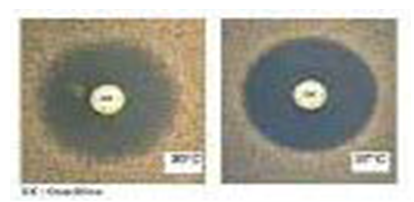 | Photo 1. Staphylococcus aureus resistant for Methicillin |
3. Results and Discussion
- Pathogens bacteria isolated from different samplesIn Total 80 strains were isolated frome urinary infections According to the results, a large dominance of Gram-negative represented by 80% were founded against 20% of Gram-positive bacteria (Figure 1).We notice that in the Gram negative bacteria, E.coli was the most frequently isolated with 78% (Figure 2). Among the Gram positive bacteria, staphylococcus aureus 5% However streptococcus 2% and 05% in different samples (Figure 2). In other Gram negative bacteria, Pseudomonas aeruginosa was isolated (Figure 2). This results was the same as reported in a study of Garraffo A and al. [11].
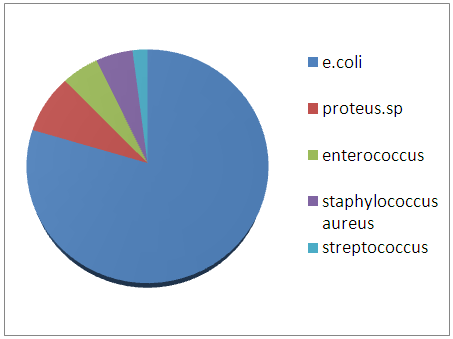 | Prevalence of strains in the urinary infections |
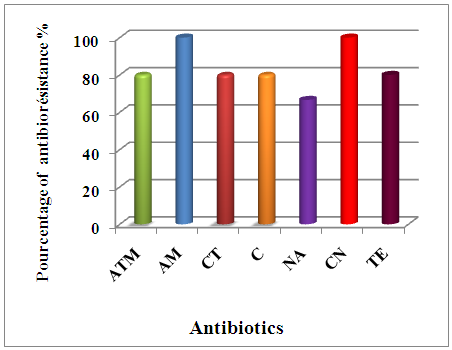 | Percentage of antimicrobial resistance of Gram-negative bacilli isolated from urinary tract infections |
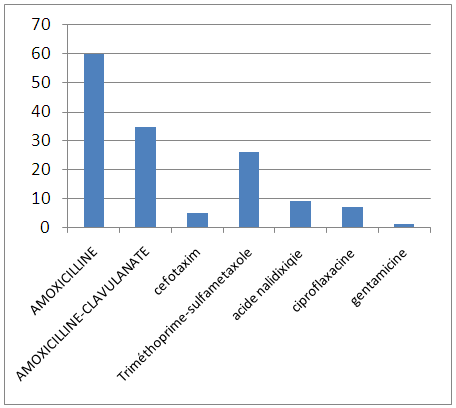 | Percentage of antimicrobial resistance of Gram-positive cocci isolated from urinary tract infections |
 Detection of beta-lactamase in Gram negative bacteriaThe result of a positive test is the inhibition zone around the discs cefixime or aztreonam in the presence of amoxicillin and clavulanic acid [16]. All strains isolated from different specimen and resistances to antibiotics were tested for the production of beta-lactamase using the spread spectrum method previously described double halo. The results indicates that the most isolated strains (79.46%) producing ESBL, which are mainly Escherichia coli, During antibiotic therapy, the acquisition of resistance determinant and the selection of resistant subpopulations in patients initially infected with a susceptible strain, presents a major problem [17]. The present study has shown that all strains are multiresistants. Previous studies have shown that ESBL-mediating plasmids may carry more than one beta-lactamase gene and that they may be responsible for high-level beta-lactamase resistance phenotypes [18]. The major risk in the beta-lactamase genes.The direct and indirect costs associated with antibiotic resistance; promote national coordination and development of action plans to respect the measures of hygiene, combat antibiotic resistance; promote the prudent use of antibiotics and the systematic implementation of infection control measures for the prevention and treatment for reduce morbidity, mortality.The use of antibiotics in human health and animal, including the impact on the food chain; review the teaching of prudent use of antibiotics in the faculties of medical sciences, veterinary and life, and implement effective policies.
Detection of beta-lactamase in Gram negative bacteriaThe result of a positive test is the inhibition zone around the discs cefixime or aztreonam in the presence of amoxicillin and clavulanic acid [16]. All strains isolated from different specimen and resistances to antibiotics were tested for the production of beta-lactamase using the spread spectrum method previously described double halo. The results indicates that the most isolated strains (79.46%) producing ESBL, which are mainly Escherichia coli, During antibiotic therapy, the acquisition of resistance determinant and the selection of resistant subpopulations in patients initially infected with a susceptible strain, presents a major problem [17]. The present study has shown that all strains are multiresistants. Previous studies have shown that ESBL-mediating plasmids may carry more than one beta-lactamase gene and that they may be responsible for high-level beta-lactamase resistance phenotypes [18]. The major risk in the beta-lactamase genes.The direct and indirect costs associated with antibiotic resistance; promote national coordination and development of action plans to respect the measures of hygiene, combat antibiotic resistance; promote the prudent use of antibiotics and the systematic implementation of infection control measures for the prevention and treatment for reduce morbidity, mortality.The use of antibiotics in human health and animal, including the impact on the food chain; review the teaching of prudent use of antibiotics in the faculties of medical sciences, veterinary and life, and implement effective policies. 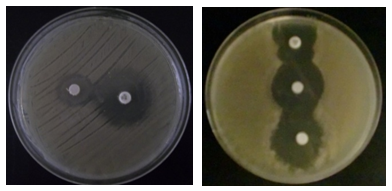 Detection of PenicillinaseOn the E. coli strains tested by the iodometric method, penicillinase-like β-lactamase was found to be 98.26%, of course after detecting discoloration which took place at least 10 min, But the rest of the strains show no visible ability to degrade penicillin G, as the initial color remains intact.
Detection of PenicillinaseOn the E. coli strains tested by the iodometric method, penicillinase-like β-lactamase was found to be 98.26%, of course after detecting discoloration which took place at least 10 min, But the rest of the strains show no visible ability to degrade penicillin G, as the initial color remains intact.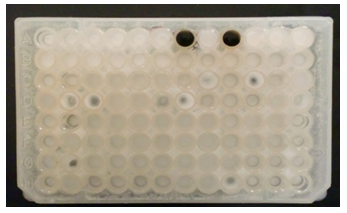 Carbapenemase DetectionThe deformation of the diameter at the intersection between a streak and the culture of Escherichia coli ATCC 25922 indicates the presence of hydrolysis of the carbapenes by the strain tested (Lee K, Chong Y., 2001).
Carbapenemase DetectionThe deformation of the diameter at the intersection between a streak and the culture of Escherichia coli ATCC 25922 indicates the presence of hydrolysis of the carbapenes by the strain tested (Lee K, Chong Y., 2001).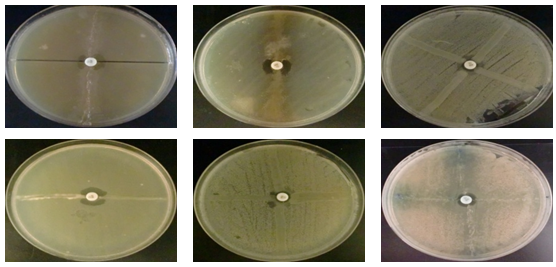 Our study indicates that most strains isolated from all three sites are producing Carbapenemases (81.81%) which is mainly produced by bacterial species that are (Escherichia coli, Klebsiella pneumoniae, Shigella sp, Enterobacter spp, Serratia marcescens and Citrobacter freundii) To 100% while the other species are variable.Beta-lactamases with carbapenemase activity are the most potent mechanisms of resistance to carbapenems. These Carbapenemases are increasingly identified in Enterobacteriaceae worldwide.However, their presence is often coupled with the presence of extended spectrum β-lactamase (ESBL), which leads to a multiresistance of secretory strains (Boutet-dubois A et al., 2012).The mechanism that can cause resistance of Enterobacteriaceae to Carbapenems: production of Carbapenemases belonging to different classes of β-lactamases (Ambler classification): class A (KPC), class B (metalloenzymes VIM, IMP, NDM) D (OXA-48, OXA-23, OXA-181 and variants thereof) (CA-SFM., 2012).In 2005, in the United Kingdom, the Health Protection Agency issued a first alert to sensitize laboratories to the emergence of carbapenemase production by enterobacteria by various mechanisms: Metallo-β-lactamases (MBL), KPC and OXA enzymes -48. Since then, the number of isolates of Enterobacteriaceae producing carbapenemases has been increasing. Isolates producing emerging metallo-β-lactamase. The bacteria involved are mainly Klebsiella pneumoniae and Escherichia coli.
Our study indicates that most strains isolated from all three sites are producing Carbapenemases (81.81%) which is mainly produced by bacterial species that are (Escherichia coli, Klebsiella pneumoniae, Shigella sp, Enterobacter spp, Serratia marcescens and Citrobacter freundii) To 100% while the other species are variable.Beta-lactamases with carbapenemase activity are the most potent mechanisms of resistance to carbapenems. These Carbapenemases are increasingly identified in Enterobacteriaceae worldwide.However, their presence is often coupled with the presence of extended spectrum β-lactamase (ESBL), which leads to a multiresistance of secretory strains (Boutet-dubois A et al., 2012).The mechanism that can cause resistance of Enterobacteriaceae to Carbapenems: production of Carbapenemases belonging to different classes of β-lactamases (Ambler classification): class A (KPC), class B (metalloenzymes VIM, IMP, NDM) D (OXA-48, OXA-23, OXA-181 and variants thereof) (CA-SFM., 2012).In 2005, in the United Kingdom, the Health Protection Agency issued a first alert to sensitize laboratories to the emergence of carbapenemase production by enterobacteria by various mechanisms: Metallo-β-lactamases (MBL), KPC and OXA enzymes -48. Since then, the number of isolates of Enterobacteriaceae producing carbapenemases has been increasing. Isolates producing emerging metallo-β-lactamase. The bacteria involved are mainly Klebsiella pneumoniae and Escherichia coli.4. Conclusions
- Our study over a short period raised significant data on the alarming development of resistance and the emergence of multidrug-resistant bacteria to antibiotics. Indeed all of the isolated bacterial strains (80) has a significant increasing trend with multidrug resistance over time as a consequence of improper use of antibiotics. The testing for beta-lactamase reveal the presence of ESBL (which reached 100% in MRSA) of penicillinase and carbapenemase in most organisms isolated. However, resistance by producing penicillinase is the type of resistance the most common. In general, and to reduce the selection pressure of antibiotics that encourages the emergence, proliferation and spread of ESBL Enterobacteriaceae, it should reduce the volume of antibiotics used in humans by intensifying actions in part of the antibiotic level. We must, in particular, introduce, next to the concept of "good use", the concept of "any use". A strong proposal to advance in this area is to define, document and make the situations in which it is recommended not to prescribe antibiotics (Rabaud Ch., 2010).
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML