-
Paper Information
- Next Paper
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
Journal of Microbiology Research
p-ISSN: 2166-5885 e-ISSN: 2166-5931
2017; 7(2): 31-38
doi:10.5923/j.microbiology.20170702.02

The Rate of Extended-Spectrum B-Lactamases among Commonly Isolated Enterobacteriaceae in Cancer Patients in Zagazig University Hospitals
Rania A. Ghonaim1, Tarek A. Elgohary2
1Departments of Clinical Pathology, Faculty of Medicine, Zagazig University, Egypt
2Department of Oncology & Haematology, Faculty of Medicine, Zagazig University, Egypt
Correspondence to: Rania A. Ghonaim, Departments of Clinical Pathology, Faculty of Medicine, Zagazig University, Egypt.
| Email: |  |
Copyright © 2017 Scientific & Academic Publishing. All Rights Reserved.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/

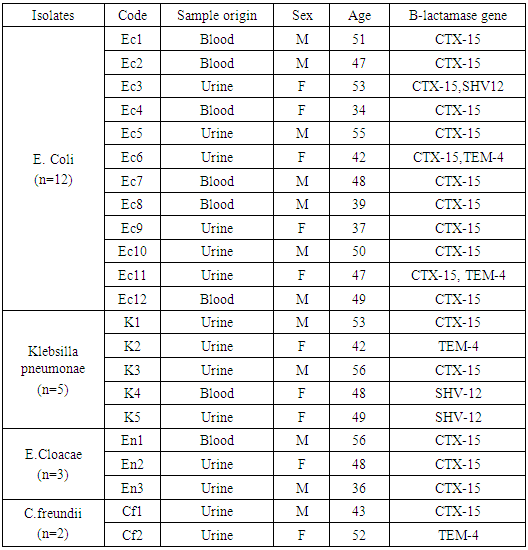
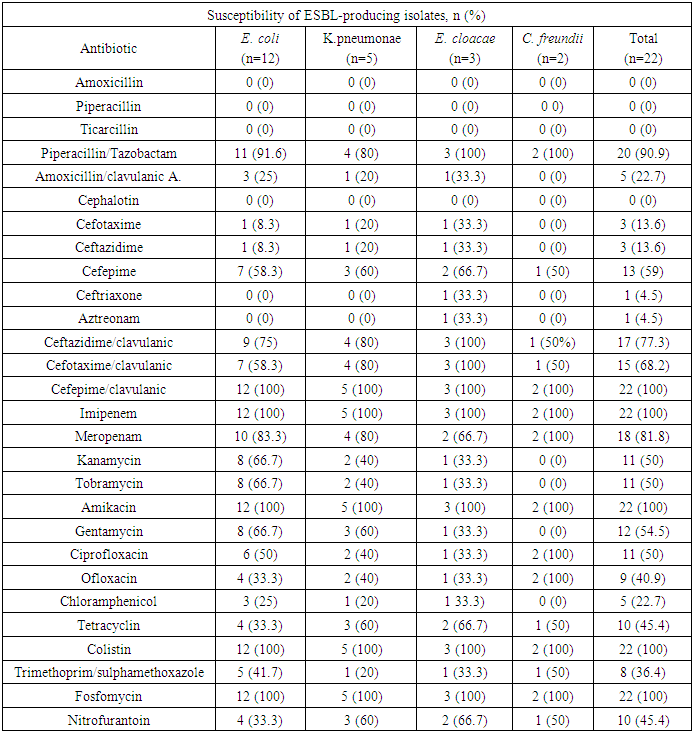
Background: The incidence of infections caused by beta-lactam resistant organisms has increased in recent years due to the production of various enzymes. Extended-spectrum β-lactamases producing Enterobacteriaceae (ESBL-E) are emerging worldwide in hospitals and in the community. Aim of the work: Thepresent study was done to determine the prevalence and molecular typing of extended-spectrum β-lactamases (ESBLs) in clinical isolates of Escherichiacoli,Klebsillapneumonae,Enterobactercloacae,and Citrobacterfreundii, isolated between January 2014 and August 2016 from the department of Oncology and Hematology, Zagazig University Hospitals. Patient & Method: Antimicrobial susceptibility testing was determined by Vitek2 & disk diffusion on Mueller Hinton agar for selection of the best antibiotic to be used in resistant cases. The characterization of ESBL genes were examined using PCR amplification and the clonal relatedness was investigated by ERIC-PCR. Results: During the study period, twenty-two (11%) isolates out of 420 specimen were found to produce ESBLs and distributed as follows: 12 isolates of E.coli(11%), 5 isolates of K.pneumonae (9.6%), 3 isolates of E.cloacae(9.37%) and 2 isolates of C.freundii(25%). ERIC-PCR was used to identify clonal relatedness, which proved that the isolates were genetically unrelated. The CTX-M-15 ESBLs were predominant representing (81.8%) of ESBL positive cases, followed by TEM-4 (18.2%) and SHV-12 (13.6%). A common band was observed in all extracts with a high molecular weight which is found to be CTX-M15 gene. The blaCTX-M-15 gene is transferred on a conjugative plasmid of high molecular weight (≈130 kb). Our results showed that all 22 strains (100%) were susceptible to imipenem, amikacin, colistin, fosfomycin and Cefepime/clavulanic Acid. Higher susceptibility levels were shown with Piperacillin/Tazobactam (90.9%) and Meropenam (81.8%). Conclusion: We conclude that there is high rate of multidrug resistant strains in our hospital with CTX-M-15 being the most common cause for Enterobacteriaceae ESBL resistance, followed by TEM-4 and SHV-12.
Keywords: Multi-resistant bacteria, Extended spectrum β-lactamase, CTX-M-15, Genotyping, ERIC PCR, (MALDI-TOF)
Cite this paper: Rania A. Ghonaim, Tarek A. Elgohary, The Rate of Extended-Spectrum B-Lactamases among Commonly Isolated Enterobacteriaceae in Cancer Patients in Zagazig University Hospitals, Journal of Microbiology Research, Vol. 7 No. 2, 2017, pp. 31-38. doi: 10.5923/j.microbiology.20170702.02.
Article Outline
1. Introduction
- Antibiotic resistance is a problem of deeply scientific importance both in hospital and community. Resistance to β-lactam antibiotics is an increasing problem and β-lactamase production is the most common mechanism of drug resistance, especially in Gram-negative bacilli. β-Lactamases are remarkably diversified due to their continuous mutation. Rapid detection in clinical laboratories is essential for the accurate recognition of antimicrobial resistant organisms.Being immunocompromised cancer patients carries the risk of infection with resistant organisms especially those receiving high dose chemotherapy. There is a high risk of neutropenic fever, so the patients′ susceptibility to be infected with resistant organisms increases. Infection by these strains of ESBL producing bacteria is associated with high mortality rate with increased risk of therapeutic failure and longer duration of hospital stay and spreading of infection consequently. Generally, ESBLs are not carried on the bacterial chromosome, however they are found on transferable element of DNA called a plasmid. Plasmid can carry many different genes and have the ability to transfer a replica of itself to other bacteria [1].Several clavulanic acid-inhibited Ambler class A expanded-spectrum β-lactamases (ESBLs) have been reported mostly in Enterobacteriaceae in addition to the TEM- and SHV-type ESBLs. One of its important types is the CTX-M-type β-lactamases, which are plasmid-mediated ESBLs and confer high level of resistance to cefotaxime, ceftriaxone, aztreonam and have only marginal effects on MIC of ceftazidime [2]. Proper infection control practices and barriers are essential to prevent the spread and outbreaks of ESBL producing bacteria. Many types of bacteria have developed different strategies to counter the effects of antibiotics. The identification of the resistance mechanism may help in the discovery & design of new antimicrobial agents. The carbapenems are widely regarded as the drugs of choice for the treatment of severe infections caused by ESBL-producing Enterobacteriaceae.
2. Patients & Methods
- This study was done in the period between January 2014 to August 2016 in both Medical Oncology & Hematology and Clinical Pathology Departments Zagazig University Hospitals. Four hundred and twenty samples were tested and only the samples showing infection by enterobacteriaceae were chosen. Identification was done on the Vitek MS (MALDI-TOF MS identification).
2.1. MALDI-TOF MS Identification
- Matrix Assisted Laser Desorption Ionization Time-of-Flight Mass Spectrometry (MALDI-TOF MS) is a rapid and inexpensive technology used nowadays for identification of most bacterial strains [3].Four hundred and twenty samples were then analyzed using the Vitek MS MALDI-TOF mass spectrometer in linear positive-ion mode, across the mass-to-charge ratio range of 2,000 to 20,000 Da. Each spot was irradiated with 500 laser shots at 50 Hz. Target plates were calibrated and quality controlled both before and after data acquisition by using Escherichia coli ATCC8739. A sample containing matrix only (negative control) were assayed for quality control purposes.After the acquisition of spectra, data were transferred from the Vitek MS acquisition to the Vitek MS analysis server and identification results were displayed using Myla v2.4 middleware. The total processing and data analysis time was approximately 20 min for a single isolate. Data Anaysis: The Vitek MS identification system is based on comparison of the characteristics of the spectra obtained with the Vitek MS v2.0 database. This database was built using spectra for known strains for each claimed species. Based on this representative data collection, a weight is assigned to each peak for each species according to its specificity. A single identification is displayed with a confidence value from 60.0 to 99.9.
2.2. Antimicrobial Susceptibility Testing and ESBL Detection
2.2.1. VITEK 2 ESBL Testing
- Each isolate was tested using the VITEK 2 system with the ESBL test panel contained in the NO45 card (all from bioMérieux). The ESBL panel has six wells containing cefepime at 1.0 μg/ml, cefotaxime at 0.5 μg/ml, and ceftazidime at 0.5 μg/ml, alone and in combination with Clavulanic Acid (CA) (10, 4, and 4 μg/ml, respectively), and growth in each well is quantitatively assessed by means of an optical scanner. The proportional reduction in growth in wells containing cephalosporin plus CA compared with those containing the cephalosporin alone is considered indicative of ESBL production and is interpreted as ESBL-positive through a computerized expert system (Advanced Expert System) [4]. Quality control strains (E. coli ATCC 25922, E. coli ATCC 35218) were included. The AST cards were filled with an inoculum (which was prepared from the 0.5 McFarland suspension and then sealed and read). The automated VITEK-2 system processed the antimicrobial susceptibility cards until the MICs were obtained [5].The susceptibility to some antibiotics, which wasn′t existed in the available vitek cards as Piperacillin/ Tazobactam, Chloramphenicol and Fosfomycin were determined by the standard disk diffusion method on Mueller-Hinton agar recommended by the Antibiogram Committee of the French Society for Microbiology [6].
2.3. Extraction of Bacterial DNA & Multiplex PCR Technique
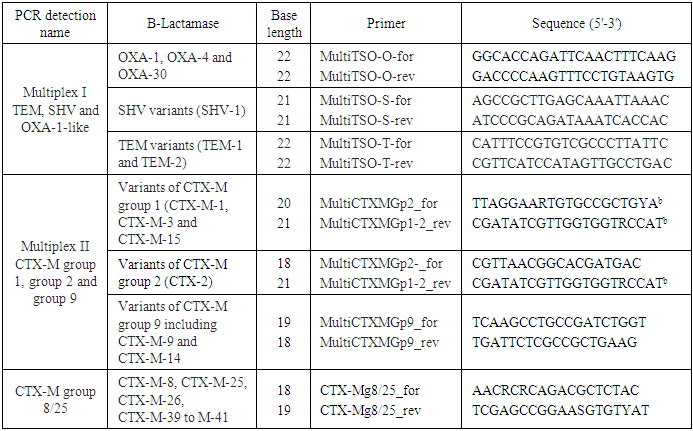
- All strains were freshly cultured on nutrient agar before DNA extctraction, which was performed by using DNA extraction kit k512 (Feremntas R) according to manufacturer instructions. Total DNA of ESBL-producing isolates was extracted by boiling to100°C for 10 min a suspension of the strains in 200 μl of distilled water and centrifugation for 7 min at 13,000 × g, then the supernatant obtained was stored at -20°C. PCR experiments were performed with these crude lysates.As many clinical isolates now harbour more than one β-lactamase and due to the high diversity of these enzymes, multiplex PCR methods are becoming widely used for their detection in epidemiological surveys [7].Multiplex PCR designed group-specific primers to amplify internal fragments of several sizes. We used two multiplex PCR in our study a blaTEM/blaSHV/blaOXA-1- multiplex PCR & a blaCTX-M multiplex PCR including phylogenetic groups 1, 2, 9, 8/25. These genotypes sequences were downloaded from the GenBank database and aligned within groups using the ClustalX software (version 2.0) as shown in (Table 3).Amplicons were visualized after running at 100V on a 2% agarose gel containing ethidium bromide for 1h. A 100 bp DNA ladder (New England Biolabs, Ipswich, MA, USA) was used as a size marker.
2.4. Enterobacterial Repetitive Intergenic Consensus PCR (ERIC-PCR)
- All ESBL producing clinical isolates were analyzed by enterobacterial repetitive intergenic consensus PCR (ERIC2-PCR). DNA was amplified using the ERIC-2 primer with the following sequence: 5'AAGTAAGTGACTGGGGTGAGCG3' [8], Amplified products were visualized on agarose gels 1.5% stained with ethidium bromide. The profiles of tested clinical isolates were considered different when at least one band differed [9]. ERIC-PCR has become a widely used technique for strain typing because of its speed and relative low cost [10].
2.5. Plasmid DNA Extraction (MiniPrep.) Using QIAGEN Kit Cat.No. 27106
- Bacteria replicate plasmid DNA before each cell division. Overnight culture of bacteria can yield vast number of plasmids [11].Ÿ Harvest bacteria by centrifuge at 13200 rpm for 2 minutes, discard supernatant. Resuspend the pelleted bacterial cells in (250 ul) resuspension buffer. Ÿ Lysis the cells by using (250 ul) of lysis buffer to release the plamid.Ÿ Neutralization buffer (350 ul) is added to precipitate cellular debris, but leaves plasmid DNA in solution.Ÿ Centrifuge to precipitate the debris.Ÿ Supernatant containing the plasmid is transferred to a spin coloumn, which contains a silica matrix. The plasmid DNA binds to silica matrix and the rest of supernatant passed through the coloumn and is discarded.Ÿ Coloumn is washed by (750 ul) washing buffer to remove any remaining contaminate.Ÿ Elution buffer is added to release plasmid DNA from silica matrix.Ÿ 50 ul of clear colourless fluid is formed on the bottom of the microcentrifuge tube containing the plasmid DNA.
3. Results
- During the study period, 200 non-duplicate clinical isolates of enterobacteriaceae were identified. Among these samples, 22 (11%) were ESBL positive by using Vitek2 AST Cards. The prevalence of ESBL production strains among these tested clinical isolates was the following: 11% (12/108) E. coli, 9.6% (5/52) K.pneumonae, 9.37% (3/32) E. cloacae and 25% (2/8) C. freundii “Figure 1’’.
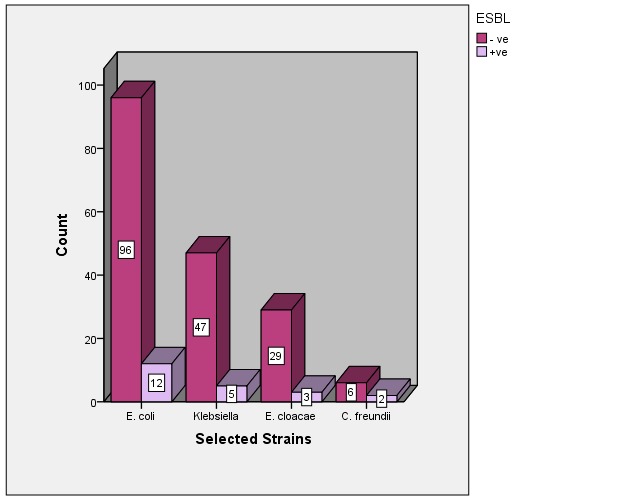 | Figure 1. ESBL +ve and ESBL –ve cases among selected strains |
|
|
|
4. Discussion
- The incidence of infections caused by beta-lactam-resistant organisms due to the production of various enzymes has rapidly increased in recent years. Extended-spectrum β-lactamases producing Enterobacteriaceae (ESBL-E) are emerging worldwide in hospitals and in the community. Their incidence varies among countries, regions or even hospitals [12].Detection of ESBL production is of great importance both in hospital and community isolates. Infection-control practitioners and clinicians need the clinical laboratory to rapidly identify and characterize different types of resistant bacteria. This is actually required to minimize the spread of these bacteria and help select appropriate antibiotics [13].Generally, ESBLs are not carried on the bacterial chromosome, but they are found on an independent element of DNA called a plasmid. Plasmids can carry many different genes on them and have the ability to transfer a replica of themselves to other bacteria. This can be very serious for many reasons. Firstly, the “other” genes could include genes conferring resistance to other classes of antibiotics that make the recipient bacteria resistant to multiple antibiotics or what is sometimes reported as a “Superbug”. Also, these plasmids have the ability to emerge in strains that do not cause human disease, but afterwards the non-pathogenic strains could transfer their plasmids to strains that can cause human disease [1].In the late 1980s, CTX-M proteins were discovered and recently more than 100 variants were known and have been sequenced. According to their aminoacid sequences, they can be divided into five groups (CTX-M groups 1, 2, 8, 9 and 25) [13].In our study, ESBLs were found in 22 (11%) of 200 clinical isolates with E. coli being the major ESBLs producer 12/22 (54.5%), followed by of K.pneumonae 5/22 (22.7%), then E.cloacae 3/22 (13.6%) and C. freundii 2/22 (9.1%). This rate of ESBLs isolates is in accordance with those reported in Kingdom of Saudi Arabia (8.9%) [14], but even higher than the rate observed in recent European studies ranging from 2.4% to 5.1% in France [15] and 2.9% in Sweden [16].In another research conducted at a tertiary hospital in Mwanza, Tanzania, the overall prevalence of ESBLs in all Gram-negative bacteria (377 clinical isolates) was 29%. The ESBL prevalence was 64% in K. pneumoniae, but 24% in E. coli [17].Recent research conducted in Ethiopia [18] reported the prevalence rate of 25% of ESBLs producers among Enterobacteriaceae family. These prevalence rates are high comparing to our study, which may be attributed to variation in drug management policies or follow other control programs.Treatment of these infections caused by ESBL-producing organisms with extended-spectrum cephalosporins or aztreonam may result in treatment failure even if the causative organisms appear to be susceptible to these antimicrobial agents by routine susceptibility testing [19]. Detection of organisms harboring ESBLs provides clinicians with helpful information. Patients colonized or infected with ESBL-producing organism should be placed under serious precautions to avoid hospital transmission [20].Our antimicrobial susceptibility testing to all ESBL-producing isolates show highly prevalent resistances against the majority of β-lactams and even to other tested antibiotics as Gentamicin, tobramycin, ofloxacin, chloramphenicols, trimethoprim and sulfonamides, which confirmed the presence of multidrug-resistant isolates in our hospital. Our results are nearly similar to the study done by Rakotonirina et al., (2013) [21].This correlates with other studies, where many ESBL producers are multi-resistant to non-β-lactam antibiotics, including fluoroquinolones and aminoglycosides [22], trimethoprim, tetracyclines, sulfonamides, and chloramphenicol, which are often encoded by the same plasmids that determine the ESBL [23].Another research was conducted at a tertiary hospital in Nigeria, among the overall ESBL producing isolates, 35% being of community origin and 65% from hospitals. The ESBL isolates showed high resistance to tetracycline, gentamicin, pefloxacin, ceftriaxone, cefuroxime, ciprofloxacin and Augmentin (Amoxicillin and clavulanic acid combination) [24].Majda et al. reported that 72% of E. coli and 65.8% of K.pneumoniae were ESBL producers at the Microbiology laboratory of Shalamar Medical College, Lahore. Sensitivity testing showed a multidrug resistance in ESBL producing E. coli and K. pneumoniae. Maximum resistance was recorded in E. coli (ESBL) as cefotaxime (98.9%), Ceftazidime (96.7%) and Cef uroxime (93.4%), while minimum resistance was seen with Imipenem (0.8%), fosfomycin (1.2%) and Nitrofurantoin as well as piperacillin/tazobactam (2.2%) each. The ESBL producing Klebsiella showed maximum resistance to ceftazidime (100%), cefotaxime (89%), and Cefuroxime (84%), while minimum resistance was seen with imipenem (4%), Nitrofurantoin and Piperacillin/Tazobactam (8%) [25]. These results comes in agreement with our results, where minimal resistance were shown with Piperacillin/Tazobactam (9.1%) and Meropenam (18.2%). Maximal susceptibility (100%) with imipenem, amikacin, colistin, fosfomycin and Cefepime/ clavulanic Acid.Molecular techniques undoubtedly have the potential to play an essential part in the laboratory setting for the screening, tracking, and monitoring of the spread of large number of organisms producing CTX-M enzymes from the community or hospital settings in optimal time.In our study, among the twenty two ESBL-producing strains isolated, CTX-M-15 was the most commonly detected genotype in all clinical isolates representing (81.8%). This comes in accordance with the Tunisian study done by [26], who reported that the CTX-M-15 ESBL gene is considered to be the most prevalent ESBL worldwide, as 32 E. coli (ESBL) isolates collected during a 10-month period. The emergence of CTX-M-15 was in 97% (31/32) of isolates and 81% (26/32) also harbored TEM-1. In recent years, there have been larger nosocomial outbreaks of clonally ESBL strains, one is recorded at a neonatal care unit with ESBL-related mortalities. A large outbreak in Uppsala involving K. pneumoniae with CTX-M-15 and in Kristiansand caused by a multi resistant CTX-M-15-producing E. coli strain [27]. According to data obtained in 2010 by the European Antimicrobial Resistance Surveillance System (EARSS), 2.6% of E. coli and 1.7% of K. pneumoniae strains in Sweden were resistant to 3rd generation cephalosporins [28].Moreover, in Madagascar, Herindrainy et al., [29] observed that 10% of non-hospitalized patients carried ESBLs, where in the majority of the cases CTX-M-15 was detected and poverty was found as a significant risk factor.In a study of inpatients in Saudi Arabia in 2008, Tawfik and colleagues found that 26% of K. pneumoniae isolates produced ESBLs, the majority of which were SHV-12 and TEM-1 enzymes, and 36% were CTX-M-15 [30].The acquisition of efficient mobile elements has accelerated the transfer of various antibiotic resistance genes. Probably, a ‘‘super bug’’, resistant to relatively all licensed antibiotics may rise in the future, so constant and careful worldwide surveillance for multidrug-resistant bacteria is urgently needed.The carbapenems (imipenem, meropenem, ertapenem, doripenem) are still the first choice of treatment for serious infections with > 98% ESBL-producing E. coli and K. pneumoniae [31], but with the emergence of the carbapenem resistant Enterobacteriaceae, which is the ‘‘magic bullet’’, It becomes difficult to be treated. There are some older drugs which can be used. Fosfomycin was reported of having admirable in vitro activity against the ESBL-producing E. coli or K. pneumoniae. In Hong Kong, most of the ESBL-producing E. coli isolates were reported to be sensitive to fosfomycin [32]. Colistin is another choice which we can consider for the treatment of these organisms. Although it is considered as quite toxic antibiotic, it is a last resort that we can consider at the present moment as there is no new antigram negative antibiotics available for the treatment of these multidrug resistant organisms [31].Careful and regular worldwide surveillance is urgently needed for multidrug resistant bacteria as superbug might exist in the future leading to resistance to relatively all licensed antibiotic.
5. Conclusions
- We conclude that there is high rate of multidrug resistant strains in our hospital with CTX-M-15 being the most common cause for Enterobacteriaceae (ESBL) resistance, followed by TEM-4 and SHV-12.
Abbreviations

 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML