-
Paper Information
- Previous Paper
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
Journal of Microbiology Research
p-ISSN: 2166-5885 e-ISSN: 2166-5931
2016; 6(4): 75-81
doi:10.5923/j.microbiology.20160604.02

Distribution of Microbial Population Associated with Penaeus monodon Larvae in Marine Nursery Ponds in Mtwapa Creek, Kenya
Mutai Edwin Kipyegon 1, Mutai Raymond 2
1Department of Biological Sciences, School of Science, University of Eldoret, Kenya
2Department of Biotechnology, School of Agriculture and Biotechnology, University of Eldoret, Kenya
Correspondence to: Mutai Edwin Kipyegon , Department of Biological Sciences, School of Science, University of Eldoret, Kenya.
| Email: |  |
Copyright © 2016 Scientific & Academic Publishing. All Rights Reserved.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/

One of the mainchallenges of prawnculture is poor survival of the larvae, which makes offspring production difficult, unreliable and expensive. The immune system of the larvae is immature and infections with pathogenic bacteria are the major cause of larval mortality. Since the larvae require life feed such as microalgae, rotifers and Artemia that need to be present in high densities, levels of dissolved and particulate nutrients are high. Due to this; saprophytic and opportunistic pathogenic bacteria thrive in cultures of marine larvae and cause infections. The present study aims to evaluate the occurrence and distribution of microbial diversity of bacterial population present in prawn larvae samples cultured in nursery ponds in Mtwapa Creek, Kenya. Microbial species were characterized based on morphological and biochemical tests. Total number of bacteria ranged from 21.7 × 105 cfu to 32 × 105 cfu. Microorganisms presumably belong to genus Vibrio, Pseudomonas, Aeromonas, Alcaligenes, Bacillus, Staphylococcus, Hafnia and Fusarium. The absence of V. harveyi pathogen indicated that the fusant serve as the main source on increasing of resistance to diseases and thus reducing the mortality of prawn larvae.
Keywords: Microbes, Pathogenic bacteria, Mtwapa, Penaeus monodon
Cite this paper: Mutai Edwin Kipyegon , Mutai Raymond , Distribution of Microbial Population Associated with Penaeus monodon Larvae in Marine Nursery Ponds in Mtwapa Creek, Kenya, Journal of Microbiology Research, Vol. 6 No. 4, 2016, pp. 75-81. doi: 10.5923/j.microbiology.20160604.02.
Article Outline
1. Introduction
- Inorganic and organic contaminants entering coastal waters may be concentrated by edible marine organisms to varying degrees from either water, their food or sediments [1]. Understanding the transfer of contaminants through the food web is critical to predict the exposure of humans to contaminants either through subsistence or commercial consumption of seafood and the possible health consequences of such exposure. In addition, such information is crucial in making accurate risk assessment for seafood safety purposes. Infectious microbial diseases and parasites are not only a major obstacle to closing lifecycles and breeding, but also inflict tremendous economic losses on the aquaculture industry. As an example, it isestimated that more than 3 billion US$ per year are lost as an effect of infectious diseases in prawn culture alone [2]. The most prevalent diseases in aquaculture are caused by bacteria (54.9%), followed by viruses (22.6%), parasites (19.4%) and fungi (3.1%) [3]. The most prevalent causative agents of bacterial infections in marine environment belong to the family Vibrionaceae of the γ proteobacteria [4]. The World’s most prominent and deleterious pathogen is Vibrio (Listonella) anguillarum. It has the ability to persist in nutrient-free sea water for more than one year and can increase a thousand fold in coastal sea water as an effect of carbohydrate-rich wastewater discharge, which may be a reason for its global abundance [5]. Vibrio harveyi has the largest impact on Crustacean and fish culture with multi drug resistant V. harveyi being a major problem in prawn culture. Outbreaks of photobacterium damselae spp. Piscicida, a member of the Vibrionaceae family, have been reported from marine aquacultures [6]. V. vulnificus and V. parahaemolyticus are of special concern, since they do not only commonly account for mortalities in aquaculture, but some strains can also cause disease in humans [7]. Pathogenic bacteria in sea water are most abundant in sediments [8] but are also seen in increased concentrations in the surface film, as compared with the water column [9]. As a result, shellfish and other benthic fish, such as Flounder, show elevated levels of these bacteria, which can also cause disease in fish, as well as human hosts [8, 10]. Many bacterial species of enteric origin can be isolated from harbours which are located around sites of human habitation, including Bacillus cereus, Staphylococcus aureus, Vibrio parahaemolyticus, Salmonella spp, Eschirichia coli, Shigellaspp, Listeria monocytogenes and Klepsiella spp. These bacterial species are commonly isolated from waters which contain fecal materials [8, 11]. The global importance of food safety is not fully appreciated by many public health authorities despite the constant increase in the prevalence of food borne illnesses. The surveillance for food borne illness has been stressed because of centralization of food production and increased International trade and tourist, the responsibility for food safety has expanded from individuals to industries and government, and thus these changes have created potentials for epidemiological outbreaks of food borne diseases. Aquatic ecosystem although harbors a sizable population of microbes [20] are often considered as an index of water quality. These ubiquitous microorganisms do find various surfaces or organs of aquatic organisms for colonization. The present study provides an account of distribution of microbial population associated with penaeus monodon larvae in marine nursery ponds in mtwapa creek, kenya.
2. Materials and Methods
- Study area and study site The Kenyan Coast is situated immediately south of the equator; it covers a distance of about 500 km while the actual length of the seafront is about 600 km. The coastline forms part of the western border of the Indian Ocean and has an almost continuous fringing coral reef. Other features of the Kenyan coast include mangrove forests and estuaries as well as a number of islands to the south, which protect several embayments and harbours [21]. Approximately three million people inhabit the Kenyan coastal areas, at a density of 300-400 persons/km2. The marine environment provides this population with employment and food in the form of shell and finfish. Fish contributes over 70% of the protein consumed by the coastal inhabitants [13]. Artisanal fishery lands 95% of the total marine catch, contributing 6% to the coastal economy, and this is the main source of livelihood for more than 60,000 households [12]. Mariculture in the Kenyan coast at the moment is still at its infancy stage. It is thus important to understand the likely ecological changes that mariculture may introduce and their remedies so that the farmers and policy makers can be guided accordingly. Facility design The study was carried out in six nursery pond units with 14m by 6m dimensions constructed at Kwetu Training Centre, Mtwapa Creek (Figure 1). The pond culture system consists of the water column and surface sediment in the ponds. The water from the creek moves through the mangrove ecosystem before getting into the ponds. The ponds were constructed in such a way that there is regulated inflow and out flow of water which is controlled by a sluice gate at the main channel entry. Water enters the ponds when the tide level rises above 3.4m but when the tide is below 3.4m no water enters the ponds. This means that the ponds are subjected to periods of no water exchange alternating with periods where there is water exchange during the high spring tide. The length of these periods varies with behavior of tides but on average water exchange takes place between 10-14 days.
 | Figure 1. Map showing the study site in Mtwapa Creek, Kenya |
3. Results and Discussion
- Bacterial Identification Larval stages of the life cycle is the most vulnerable to prawn disease. The present study deals with the distribution of bacteria which are suspected to be the major reason in causing mortality of P. monodon larvae in Nurseries. The total numbers of isolated bacteria were 21.7 × 106 (approx. 5.4 × 104/g of larval macerate) from 2000 samples of prawn larvae as shown in table 1 and figure 2. This bacterial number was very important to note since one of the first criteria of microbiological test for evaluating prawnlarvae quality is that the maximum total bacteria count should be 1.0 ×103 CFU/g of larval macerate in agar, of which more than 90% of the colonies should be yellow [17]. The study shows that the occurrence of total bacteria in P. monodon larvae exceeded the allowed maximum number and therefore the mortality of larvae was mainly due to bacteria. Furthermore, most of the colony on NA plate had a yellow colour, instead of white and pale which could have been possibly due to presence of suspected several genus of Vibrio and its related genera consisting of Aeromonas Hafnia, Pseudomonas and Alcaligenes. Research has shown that Vibrio strain are pathogenic and can cause Vibriosis, a serious infectious disease in maricultured organisms [18]. Several Vibrios associated with shrimp larvae, juvenile and adult stages consist of V. alginilyticus, V. parahaemolyticus, Photobacterium damselae, and V. mimicus [19].
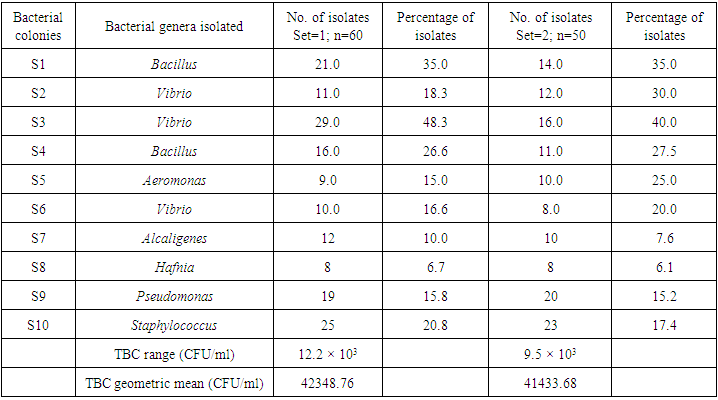 | Table 1. The number and percentage of bacterial isolates from Penaeus monodon larvae (set=1; n=1000 and set=2; n=1000) |
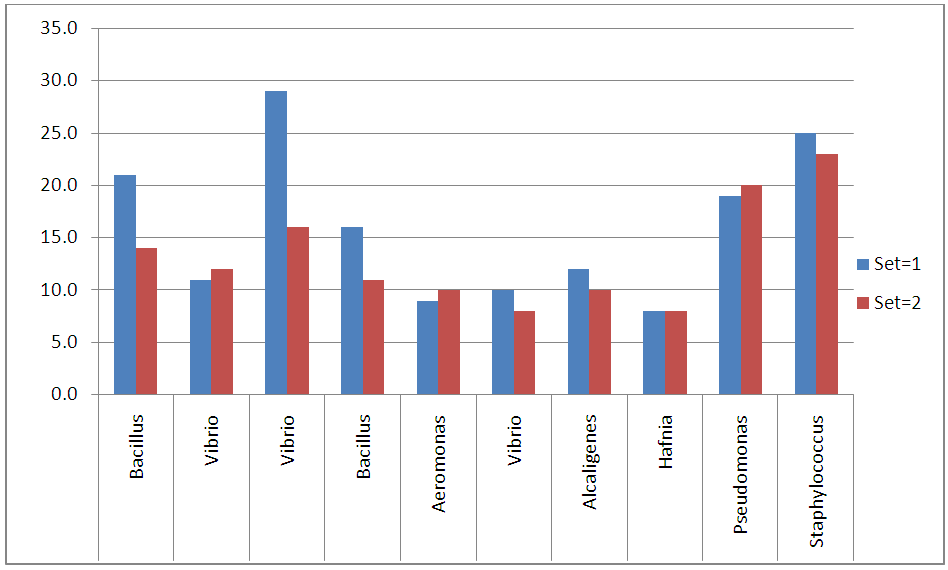 | Figure 2. The number and percentage of bacterial isolates from Penaeus monodon larvae (set=1; n=1000 and set=2; n=1000) |
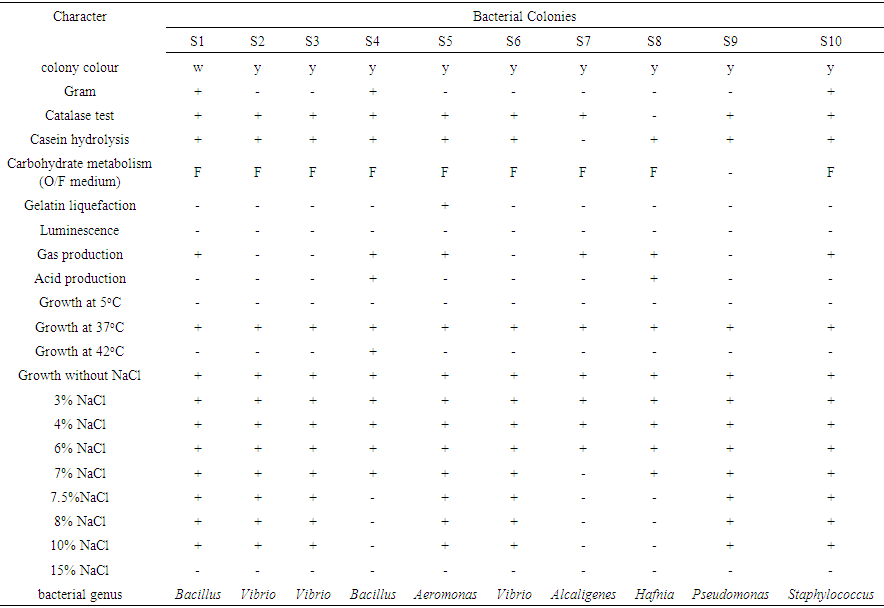 | Table 2. Characteristics of the genus of selected bacteria |
4. Conclusions
- Total number of bacteria ranged from 21.7 × 105cfu to 32 × 105cfu. Microorganisms presumably belong to genus Vibrio, Pseudomonas, Aeromonas, Alcaligenes, Bacillus, Staphylococcus, Hafnia and Fusarium. The absence of V. harveyi pathogen indicated that the fusant serve as the main source on increasing of resistance to diseases and thus reducing the mortality of prawn larvae. The study hasalso indicated the possibility of microorganisms on prawn larvae as probiotic which remain to be explored.
ACKNOWLEDGMENTS
- We are grateful to the National Council of Science and Technology (NCST) for funding the project and Kwetu Training Centre for hosting the project. We thank Mr. Brendan Muli for technical assistance in the field during the study.
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML