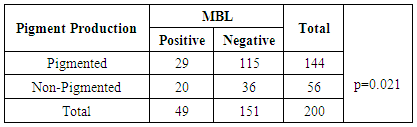-
Paper Information
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
Journal of Microbiology Research
p-ISSN: 2166-5885 e-ISSN: 2166-5931
2015; 5(6): 175-180
doi:10.5923/j.microbiology.20150506.02

Detection of blaIMP Gene in Metallo-β-Lactamase Producing Isolates of Imipenem Resistant Pseudomonas aeruginosa; an Alarming Threat
Sidra Shan1, Saira Sajid2, Khawaja Ahmad3
1Department of Microbiology, Madinah Teaching Hospital, Faisalabad, Pakistan
2Department of Pathology, University of Nottingham, United Kingdom
3Department of Biotechnology, The University of Punjab, Lahore, Pakistan
Correspondence to: Sidra Shan, Department of Microbiology, Madinah Teaching Hospital, Faisalabad, Pakistan.
| Email: |  |
Copyright © 2015 Scientific & Academic Publishing. All Rights Reserved.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/

Pseudomonas aeruginosa is one of the major cause of infections. There are two mechanisms by which P. aeruginosa has developed resistance against imipenem: by decreased drug permeability through the outer membrane proteins known as OprD and metallo-β-lactamase (MBL) production. Approximately 9 different types of MBL genes have been identified. Among them, blaIMP is the most prevalent one. Collected 200 laboratory isolates subjected for antimicrobial testing. Among them 28% (n =56) P. aeruginosa isolates were resistant to imipenem known as Imipenem Resistant P. aeruginosa (IRPA). Among all IRPA isolates; 87.5% (n=49) were found to be MBL producers. The detection of blaIMP gene was done after by ribotyping of IRPA strains. No blaIMP gene detection was present in all these MBL positive P. aeruginosa.
Keywords: P. aeruginosa, IRPA, MBL, blaIMP gene
Cite this paper: Sidra Shan, Saira Sajid, Khawaja Ahmad, Detection of blaIMP Gene in Metallo-β-Lactamase Producing Isolates of Imipenem Resistant Pseudomonas aeruginosa; an Alarming Threat, Journal of Microbiology Research, Vol. 5 No. 6, 2015, pp. 175-180. doi: 10.5923/j.microbiology.20150506.02.
Article Outline
1. Introduction
- Infectious diseases have affected the health of humans in many aspects and always remained one of the major causes of mortality and morbidity [1]. Amongst different microbial pathogens, Pseudomonas aeruginosa has been declared as one of the major cause of infectious agents belonging to the group of gram negative bacteria. It’s an alarming threat to the hospitalized and immunosuppressed patients and has the ability to persist in different environmental conditions [2]. According to Center of Disease Control and Prevention (CDC), P. aeruginosa has been ranked as the fourth most commonly isolated nosocomial pathogen and its multidrug resistance poses a challenge for its treatment [3]. The emergence of imipenem resistance in P. aeruginosa has become a severe threat, especially in poor and developing countries due to the limitation of sources including Pakistan. The epidemiological data available in Pakistan indicates that the P. aeruginosa is one of the frequent pathogen cause infections in hospitalized patients. Other reports in Pakistan reported that in clinical isolates of P. aeruginosa an increase in percent resistance against imipenem was observed [4, 5]. Carbapenems and fluoroquinolones are potent and first line of therapy against P. aeruginosa. But now in some reported cases, P. aeruginosa has developed resistance against fluoroquinolones [6]. Carbapenems are the second choice of treatment and have been developed from thienamycin, a naturally derived product of Streptomyces cattleya. They inhibit L-D-transpeptidases during the cell wall synthesis. Among carbapenems known; imipenem is more important and effective antibiotic. It is the first-line of therapy against pseudomonal infections. It can bind strongly with penicillin binding proteins (PBPs), especially with PBP-2, PBP-1a and PBP-1b [7]. Resistance against imipenem knows as Imipenem Resistant P. aeruginosa (IRPA). There are two mechanisms by which P. aeruginosa has developed resistance against imipenem: by decreased drug permeability through the outer membrane proteins known as OprD and carbapenem hydrolyzing β-lactamase (metallo-β-lactamase). The resistance may be either by one or both of the two mechanisms [8]. The second major cause other is the metallo-β-lactamase (MBL) production. Metallo-β- Lactamase (MBL), according to the Ambler's classification is class B. It’s a Zn2+ dependent carbapenem, active site of it have zinc ions for nucleophilic attack by zinc-bound water hydroxide. It is resistant to clavulanic acid but can be inhibited by EDTA (Ethylene-Diamine-Tetra-Acetic Acid), Tetramethylethylenediamine and other chelating agents [9].MBL was first detected in Bacillus cereus, in 1960s and they have intrinsic chromosome resistance against β-lactam antibiotics. Approximately 9 different types of MBL have been identified in gram negatives but most common and important types of acquired MBLs include IMP (Imipenem Carbapenemase) (Kyorin Health Science MBL 1) is rare [10, 11]. In 1991, first plasmid-mediated MBL gene, IMP-1 was identified in P. aeruginosa from Japan [4], then subsequently other IMP-type of MBLs were detected in other Eastern countries, including Europe, Canada and Brazil. Analysis of gene blaIMP showed the features of different class integrons. Integrons are DNA-based structures that carry the genes of antibiotic resistance, known as gene cassettes and most transferable MBLs are encoded by type 1 and 3 integrons [12, 13]. Objectives of Study: Main objective of this research is to investigate the prevalence of Imipenem Resistant P. aeruginosa (IRPA) and detection of blaIMP gene in the confirmed cases of MBL strains.
2. Material and Methods
2.1. Study and Location
- Our study was carried out in the Government College University, Lahore and King Edward Medical College Lahore.
2.2. Bacterial Isolation and Identification
- Two Hundred laboratory samples were collected and inoculated on MacConkey and Blood agar for the isolation of P. aeruginosa. Initial identification was done phenotypicaly including API 20NE galleries (BioMerieux, France).
2.2.1. Antibiotic Sensitivity Test and Detection of Imipenem Resistance
- Antimicrobial sensitivity testing was performed by according to CLSI recommendation 2012, in order to investigate the presence of imipenem resistance in P. aeruginosa.
2.2.2. Detection of MBL
- IRPA are MBL producers. Confirmation of metallo-β- lactamase production was done by imipenem-EDTA combined disk diffusion method. MBL need Zn2+ at its active site for activation. Whereas, EDTA as a chelating agent to inactivate the MBL by capturing the zinc ions. EDTA solution of 0.5 M (750 µg EDTA) concentration was prepared in this method by adding 186.1 g of di-sodium EDTA. 2H20 (Sigma) in 1 liter distilled water. NaOH was added to adjust the pH of solution 8.0. This solution was sterilized by autoclaving. On the dried plate of Mueller-Hinton agar two imipenem (10 µg) disks (Oxoid, UK) were placed. Out of two imipenem disks, on one disk 4 µl volume of 0.5 M EDTA was added. Plates were incubated for 18-20 hours at 35°C. EDTA causes an increase in the zone of inhibition from 8-15 mm (imipenem plus EDTA) were regarded as MBL producers, whereas, an increase in the zone of inhibition from 0-5 mm was regarded as MBL non-producers [14] (as shown in the Fig. 1).Positive and negative controls as P. aeruginosa (MBL positive) and P. aeruginosa (MBL negative) respectively were also included.
2.2.3. Ribotyping
- All the imipenem resistant MBL positive P. aeruginosa isolates were re-confirmed by ribotyping of 16S rRNA by polymerase chain reaction (PCR) and amplified product was isolated on 1% agarose gel electrophoresis. The forward and reverse primers are utilized for PCR reaction was as follows [15]:Forward Primer: 5' -AGA GTT TGA TCC TGG CTC AG -3' Reverse Primer: 5' –TAC GGT TAC CTT GTT ACG ACT T- 3'The correct size of end product and for the separation of mixed population of DNA was determined by running the product on 1% agarose gel.
2.2.4. Phylogenetic Analysis
- Phylogenetic trees, based on homology search results through BLAST [16] were constructed taking the 16S rRNA sequences of each of IRPA sequence as query. As the 16S RNA is very much conserved in all of the bacteria, therefore, during BLAST search the option of ‘similar sequences’ was chosen instead that of ‘dissimilar sequences’ so as search may include more diverse sequences to find the Phylogenetic relationship of 16S RNA sequence of IRPA isolates of this study. Moreover, 16S ribosomal RNA sequences database was selected for the homology search through BLAST at NCBI. The Phylogenetic trees of relationship were through neighbor joining method using percent identity through Jalview [17].
2.2.5. Detection of blaIMP Gene
- For the detection of blaIMP gene, PCR and 1% agarose gel electrophoresis was performed on MBL producing isolates of P. aeruginosa. The primers utilized for PCR reaction were as follows [18].blaIMP Forward Primer: 5' - GAA GGY GTT TAT GTT CAT AC-3' blaIMP Reverse Primer: 5' - GTA MGT TTC AAG AGT GAT GC - 3'The correct size of end product and for the separation of mixed population of DNA was determined by running the product on 1% agarose gel.
2.2.6. ATCC Strain
- ATCC 27853 Pseudomonas aeruginosa and were included to monitor quality control.
2.2.7. Statistical Analysis
- Data was analyzed by using computer software program SPSS 19.0. The values are expressed in percentages. Chi square test was used for the determination of the statistical significance (p-value).
3. Results and Discussion
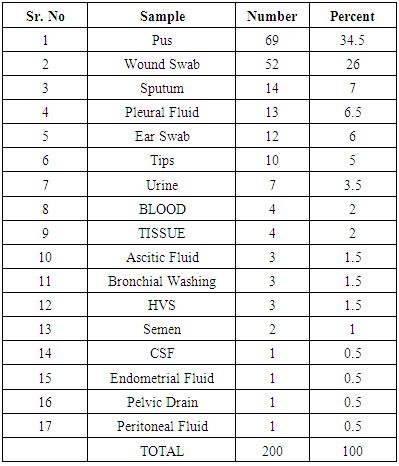
- In the present study, we have collected 17 different categories of clinical samples (Table 1). The percentage of the indoor patients 70.5% (n=141), having P. aeruginosa, were observed to be much higher compared to the outdoor patients 29.5% (n=59). These findings are similar to the study of Prashant et al., in 2011 [19]. According to them, indoor patients, with P. aeruginosa, were 84.92% compared to the outdoor patients (15.07%). These results correspond to the fact that duration of hospital stay has been related with incidence and prevalence of P. aeruginosa infections.
|
|
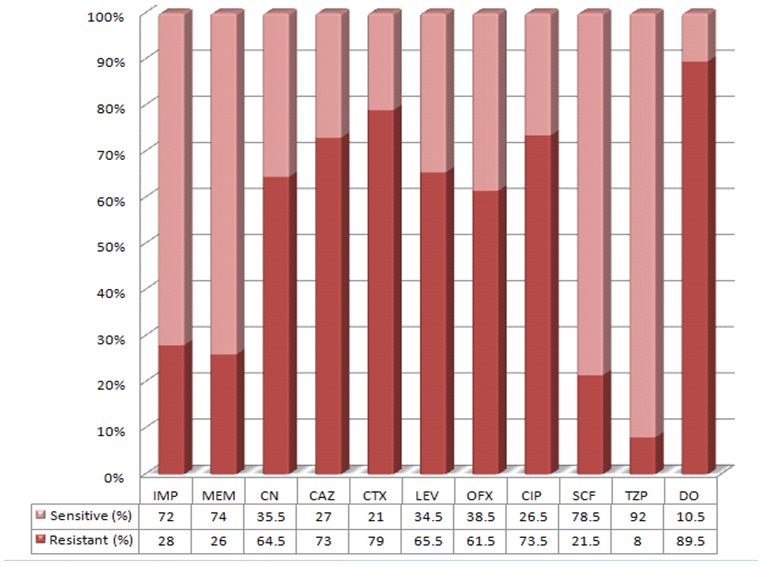 | Figure 1. Resistance Pattern of 200 isolates of P. aeruginosa against various antimicrobials (%age) |
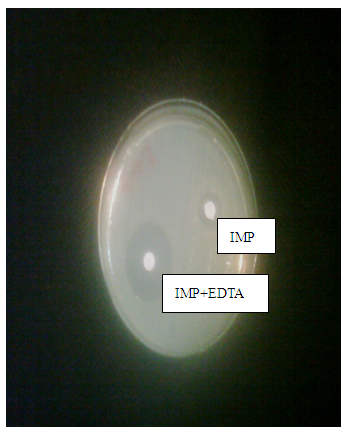 | Figure 2. MBL Detection by Imipenem-EDTA Disc diffusion Method |
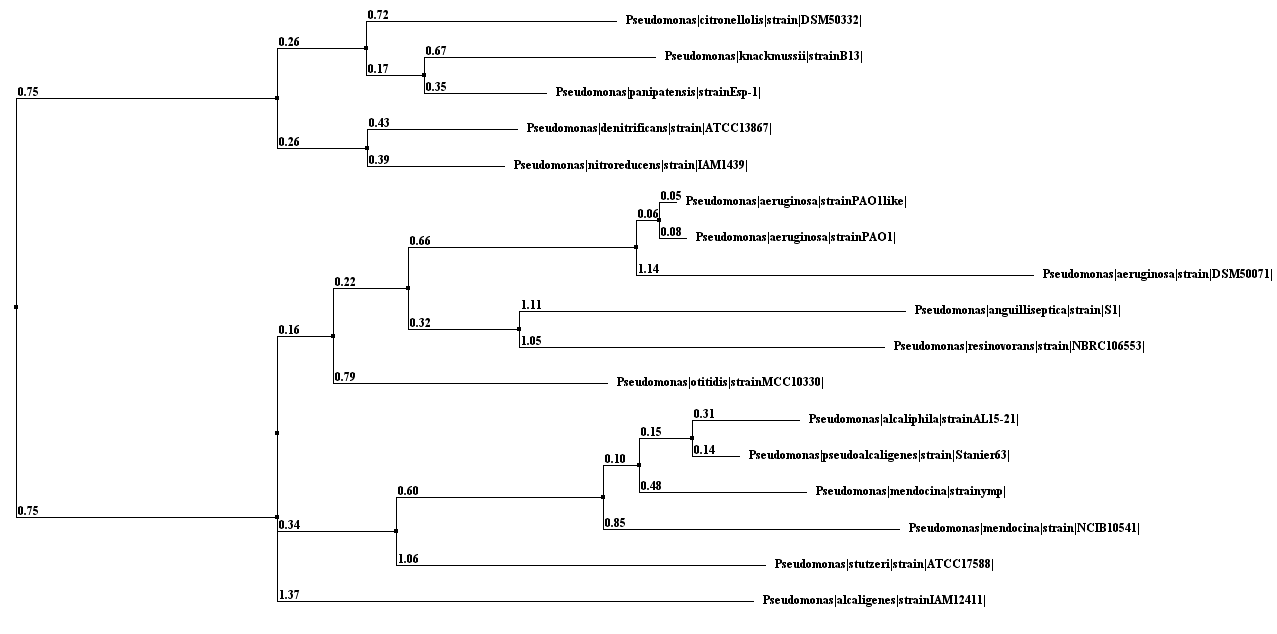 | Figure 3. Phylogenetic Tree |
 | Figure 4. 1Kb DNA Ladder, 2-4: Test isolates for blaIMP detection |
4. Conclusions
- Overall, 28% IRPA has been observed in this study which is an important issue regarding the treatment of infection. Drugs including Tazocin and Sulzone have been found to be the most effective antimicrobials against IRPA. However no blaIMP gene has been found in our hospital settings but still scope of detection for the presence of MBL gene in Pakistani IRPA isolates is much high.
ACKNOWLEDGEMENTS
- We are grateful to kind Edward Medical College, Mue Hospital Lahore for providing us the clinical materials for analysis and with special thanks to the University of Punjab Lahore, for the use of their research facilities and to all the participants.
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML