-
Paper Information
- Next Paper
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
Journal of Microbiology Research
p-ISSN: 2166-5885 e-ISSN: 2166-5931
2015; 5(3): 77-83
doi:10.5923/j.microbiology.20150503.01
Genetic Expression of MecA Gene in Methicillin Resistant Staphyococcus aureus (MRSA) Strains of Animal and Human Samples
Ghada T. Y. Helal1, Mona I. El-Enbaawy2, Soaad A. Nasef3
1Modern Veterinary Lab –General organization for veterinary service (G.O.V.S.), Fayoum, Egypt
2Professor of Microbiology, Faculty of Veterinary Medicine, Cairo University
3Chief Researcher of Poultry diseases (N.Q.P.-A.H.R.I)
Correspondence to: Ghada T. Y. Helal, Modern Veterinary Lab –General organization for veterinary service (G.O.V.S.), Fayoum, Egypt.
| Email: |  |
Copyright © 2015 Scientific & Academic Publishing. All Rights Reserved.
It is important to understand the zoonosis ,incidence and the relation between Staphylococcus aureus infection especially Methicillin Resistant Staphylococcus aureus (MRSA) in human and animal in Egypt and its public health hazards.Samples from human and animal origin suspected to have Staphylococcus aureusinfections were collected from Al Fayoum, Giza, BeniSuef and Cairo Governorates under complete aseptic conditions. Isolation and identification of Staphylococcus aureususing standard methods, antibiogramtesting to select the multidrug resistance strains and detection of mecAgene and hlggene in multidrug resistance strains by polymerase chain reaction (PCR) were done. Results showed that, 91.9℅ of the human nasal swabs were positive for Staphylococcus aureuswhile only 8℅ of them revealed non Staphylococcus aureusisolates. All animal samples were positive for Staphylococcus aureusexcept 21 cases of poultry chronic respiratory disease (CRD) where all of them have microorganisms other than Staphylococcus 38 (66.6℅) of total 57 human nasal swabs were Methicillin Resistant Staphylococcus aureus (MRSA)and only 19 samples (33.3℅) were non-Methicillin Resistant Staphylococcus aureus (MRSA). All sheep abscess pus and bumble foot samples were MRSA. While, 7 (77.7℅) out of 9 mastitic milk samples were Methicillin Resistant Staphyococcus aureus (MRSA) and only 2 (22.2℅) of these 9 samples were non-Methicillin Resistant Staphyococcus aureus(MRSA). All Methicillin Resistant Staphylococcus aureus (MRSA) isolates either from human or animal origin were mec A gene positive by PCR. On the other hand, only 80℅ of non-Methicillin Resistant Staphyococcus aureus (MRSA) isolates of human origin were positive for mecA gene by (PCR). The highest percent of hlg gene PCR positive results were represented in Methicillin Resistant Staphylococcus aureus (MRSA) isolates of animal origin 80℅. Methicillin Resistant Staphylococcus aureus (MRSA)isolates of human origin with hlg gene PCR were 60℅ which is higher than those non-Methicillin Resistant Staphylococcus aureus(MRSA) isolates of human origin which was 40℅.FourStaphylococcus aureusisolates of human origin were non-Methicillin Resistant Staphylococcus aureus (MRSA) by disc diffusion and they were positive for mec A by PCR. The PCR results and the results of disc diffusion method were correlated in 11 isolates out of 15 animal and human isolates. The strains isolated from human and animal that showed haemolysis on sheep blood agar were positive for hlg gene by PCR.
Keywords: MRSA, PCR, mecA gene, Human, Animals
Cite this paper: Ghada T. Y. Helal, Mona I. El-Enbaawy, Soaad A. Nasef, Genetic Expression of MecA Gene in Methicillin Resistant Staphyococcus aureus (MRSA) Strains of Animal and Human Samples, Journal of Microbiology Research, Vol. 5 No. 3, 2015, pp. 77-83. doi: 10.5923/j.microbiology.20150503.01.
1. Introduction
- S. aureus could cause mastitis in cows, sheep and goats, leading to severe economic losses worldwide [1, 2].MRSA has been found to colonize live stock including pigs, cattle and poultry. Since many of the MRSA clonallineages identified in livestock were un-Common for MRSA isolates found until then in human hosts, the term ‘‘livestock-associated MRSA’’ (LA-MRSA) has been introduced to distinguish these MRSA from classical human hospital-acquired (HA-MRSA) or community-associated MRSA (CA-MRSA) [3].A part from having pathogenic versatility, S. aureus can adapt rapidly to the selective pressure of antibiotics, with the emergence and spread of methicillin-resistant S.aureus (MRSA) isolates being a relevant example. MRSA was first described in 1961, the year in which methicillin was marketed [4], and actually most of the nosocomial S. aureus infections are caused by methicillin-resistant S.aureus strains [5], which have become a widely recognized cause of morbidity and mortality throughout the world [6]. S. aureus becomes methicillin resistant by the acquisition of the mecA gene which encodes a penicillin binding protein (PBP2a) with a low affinity for β-lactamas. The strains producing PBP2a are resistant to all β-lactams [7]. Thus, MRSA strains resistant to quinolones or multiresistant to other antibiotics have been emerging, leaving a limited choice for their control [8].So, the aim of this work is the characterization of MRSA strains from human and animal phynotypically and genotypically. Hemolysin gene was used as an indicator for Stapylcoccus aureus by PCR and mec A gene was used as an indicator for MRSA.
2. Methods
- Sample collectionThis study was carried on 100 samples from human and animal (poultry, cow and sheep) source. The samples were collected through 2013-2014. They were collected from Al Fayoum, Giza, BeniSuef and Cairo Governerates. Human samples were from nasal swabs of respiratory infected patients, bumble foot swabs, caseated material from poultry CRD. Mastitic milk samples and sheep swab from pus of abscess were collected.• Isolation of S. aureus was done according to standard methods [9].• Identification of isolated Staphylococci was done according to standard methods [9].• Sensitivity of isolated S. aureus was done according to standard methods [9].From the 100 isolates including human and animal samples, representative samples will be exposed to PCR for mecA gene and hlg gene presence confirmation in the central laboratory for veterinary quality control on poultry production in Dokki.Extraction of DNA was done according to QIAamp DNA mini kit instructions. Preparation of PCR Master Mix for PCR according to Emerald Amp GT PCR mastermix (Takara) Code No. RR310Akit. Cycling conditions of mecA primers during PCR was done according to standard methods [10]. Cycling conditions of hlg primers during PCR was performed according to standard methods [11]. Agarose gel electrophoreses was done according to standard methods [12].
3. Results
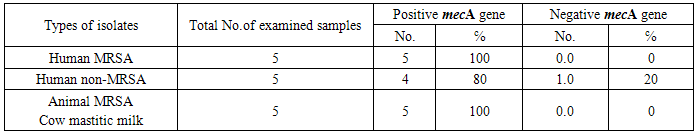
- S.aureus isolated from different animal samples are higher than in human samples. While Staphylococcus spp. other than S.aureus are higher in human samples than in animal samples. That’s to say, 91.9℅ of the human nasal swabs were positive for S.aureus while only 8℅ of them reveled non S.aureus isolates and all animal samples were S.aureus except for the 21 case of poultry CRD where all of them were negative. It was also found that MRSA isolates were higher in animal samples than in human samples. non-MRSA isolates were higher in human samples than in animal samples.38 (66.6℅) of total 57 human nasal swabs were MRSA and only19 samples (33.3℅) were non-MRSA. All sheep abscess pus and bumble foot samples were MRSA. While, 7 (77.7℅) out of, 9 mastitic milk samples were MRSA and only, 2 (22.2℅) of these 9 samples were non-MRSA.All S. aureus isolates either from animal or human origin were found to be haemolytic on sheep blood agar.Table (1) shows that all MRSA isolates either from human or animal origin were mecA gene positive by PCR. On the other hand, only 80℅ of non-MRSA isolates of human origin were positive for mecA gene by PCR. Photo (1) illustrated a 391 bp for mec Agene in positive samples from animal and human. Sample in lane 4 was negative for mecA gene by PCR.
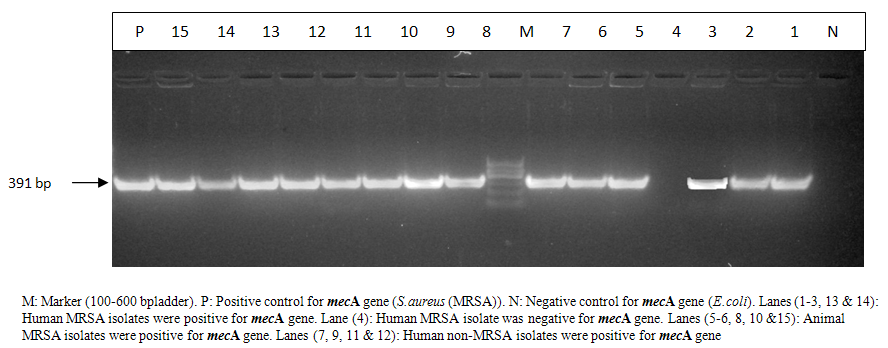 | Photo (1). Electrophoretic pattern of mecA gene in MRSA strains isolated from animal and human samples |
|
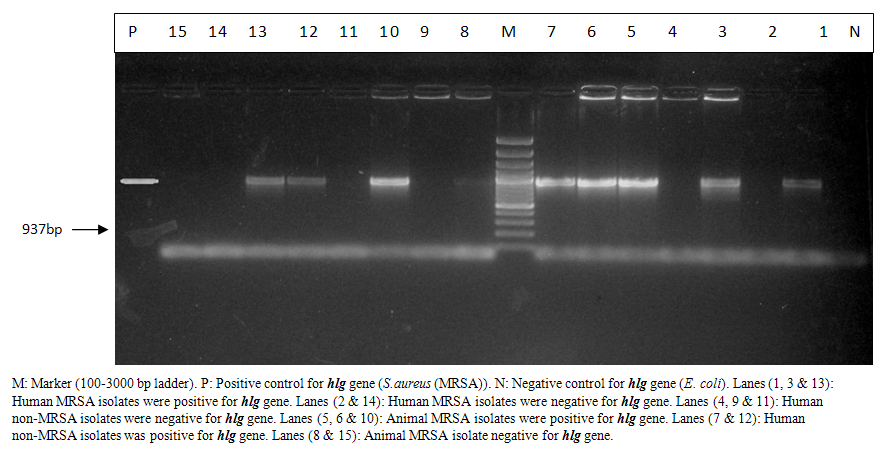 | Photo (2). Electrophoretic pattern of hlggene of MRSA isolated from animal and human samples |
|
4. Discussion
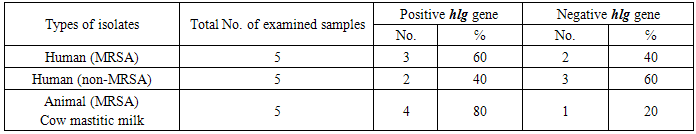
- The primary habitat of S. aureus is in the nasal passage on the skin and hair of human and warm-blooded animals. The transmission of the organisms may occur through skin lesions, contaminated food, including milk and other animal products [13]. S. aureus also could cause mastitis in cows, sheep and goats, leading to severe economic losses worldwide [2]. In humans, this bacteria causes food poisoning, toxic shock and variety of pyogenic infections [14].Concerning the human samples included in our study, 57(91.9℅) isolates out of 62 Staphylococcus isolates were S. aureus while only 5 (8℅) isolates were non S. aureus. These results disagreed with Habeeb et al. [15] who detected S.aureus in 90 (18.4%) of 489 students carried S.aureus.S. aureus was isolated from nasal swabs of 102 (40.8%) out of the 250 volunteers in Brazil [16]. In northern Pakistan, the nasal colonization of S. Aureus was documented in 86 out of 360 students (24%) [17]. In the United States from 2003-2004, the S. aureus carriage rate in the civilian non-institutionalized population according to the National Health and Nutrition Examination Survey was 28.6% [18]. S.aureus was isolatedin a low percentin India as the nasal carriage rate of S. aureus was 13% and it was only 12.6% in Sudan [19], and Onanuga and Temedie [20] found that 40 (33.3%) S. aureus strains were isolated from the nasal swabs screened in Nigeria.Resistance to several drugs was determined by plating on trypticase soy agar containing antibiotics. After 24h incubation, growth of more than two colonies was determined as resistance. In addition, resistance to meticillin was detected on oxacillin resistance screening agar (Mueller–Hinton agar+oxacillin) and was confirmed by screening forpenicillin-binding protein 2a (Slidex MRSA detection; Denka Seiken) [21]. In the present study, 57 Human nasal swabs showed 38 isolates (66.6℅) of MRSA strains. This result disagreed with Habeeb et al. [15] who found that only 10 (2.04%) of the students were found to be MRSA carrier and he exceeded that the nasal carriage of MRSA among the S. aureus isolates was 11.1% [15]. In Palestine in 2011, it was found that the nasal carriage of MRSA among the students was 2.5% [17]. In 2010, a total of 322 university students in Taiwan were screened and 2.2%, of them harboured MRSA [22]. In Pakistan, from 2007 to 2008, MRSA isolated from nasal swabs from anterior nares was 1.5% [23]. In 2001-2002, it was reported that national MRSA colonization prevalence in USA was 0.8% [24].There have been few investigations of MRSA in poultry. Two reports describe MRSA isolation from healthy and sick chickens [25, 26], but there are limited prevalence and incidence data. Two recent studies reported isolation of sequence type 398 (ST398) from healthy chickens [27, 28] and a third, involving characterized isolates from infected ultry, reported the predominance of a common human epidemic clone (clonal complex 5) [29].To determine whether MRSA is present in poultry, 50 laying hens and 75 broiler chickens were examined. MRSA was found in some broiler chickens but no laying hens. In all samples, spa type t 1456 was found [30].Youssef and Hamed [31] performed a study as they confirmed that to the best of our knowledge Staphylococcosis caused by S. aureus impacts on chicken broilers. Its public health hazards have not been illustrated in Egypt. As their study aimed to estimate the incidence, antibiotic resistance profile and zoonotic implications of S. aureus (MRSA) related arthritis in some broiler farms where they took samples from birds with clinical findings of depression and inability to stand, arthritis with swelling and local worming at hock and stifle joints and necropsy showed whitish to yellowish exudates at affected joints were collected from 20 broiler farms at finishing ages (>30 days) located at Ismailia governorate, Egypt. Swabs from joint exudates were tested for S. aureus on the basis of cultural and biochemical properties and confirmed by PCR amplification of 16S rRNA gene. Results showed that, 13/20 (65%) farms, 110/200 (55%) arthritic birds, 7/60 (11.7%) apparently health and 7/20 (35%) litter samples were positive for S. aureus. Coagulase positive strains were isolated from 11 (65%), 93(46.5%), 5 (8.3%) and 6 (30%). The in vitro antibiotic sensitivity test revealed that 58.2% of the isolates were completely resistant, 35.3% were moderately sensitive and 6.5% were highly sensitive to 17 different antibiotic discs. Complete antibiotic resistance to methecillin and cloxacillin, oxytetracycline and Sulbactin- Ampicillin were observed in all isolates.In farm workers, 14 (31.1%) of 45 were S.aureus carriers. All human isolates were multi-drug resistant (MRSA) strains and none of the workers had skin affection. In conclusion, MRSA infections were prevalent among broilers at the finishing ages; it was a potential cause of economic losses by arthritis and posing a health hazard of zoonotic transmission to human contacts and consumers [31].In the present study, we could detect S. aureus isolates among 15 animal samples out of 100 examined samples, out of which 9 samples were mastitic milk due to S. aureus infection. The rest of animal isolates were 3 isolates from sheep abscess and 3 isolates from bumble foot of poultry. The rest of the 21 isolates from poultry CRD were microorganism other than Staphylococcus. From the isolated S.aureus,13 isolates from animal samples represented by 86.6℅ out of which 3 isolates (100℅) from bumble foot of chicken and 3 isolates (100℅) were from sheep pus from abscess were methicillin resistant by using disk diffusion technique.Concerning animal samples, all sheep abscess pus and bumble foot samples were MRSA. While, 7(77.7℅) out of 9 mastitic milk samples were MRSA and only 2(22.2℅) of these 9 samples were non–MRSA isolates. S. aureusis one of the most important bacterial pathogens in bovine mastitis, a disease that causes significant economical losses in the milk industry; thus, S. aureus in general and MRSA in particular have been the focus of several studies in dairy cattle. Devriese and Hommez [32], were the first to report MRSA in bovine mastitis milk comes from Belgium where in 1972 where isolated strains that, using biotyping methods, appeared to be of human origin.On the other hand, in the present study, 7(77.8) MRSA isolates out of total 9 S.aureus isolates were from cow mastitic milk. While, only 2 (22.2℅) S.aureus isolates showed non-MRSA isolates. A study in the Republic of Korea was performed and isolated a small proportion (0.4%) of MRSA among 3047 bacterial isolates from bovine mastitis milk [33]. Alsoin Switzerl and in Japan, MRSA was isolated only from 1.4% and 1.10℅ of mastitic milk samples, respectively [34, 35]. Regarding human samples, 57 isolates out of 62 examined samples were suspected MRSA by using disc diffusion technique where our results agreed with Rushdy et al. [36] who isolated out of 200bacterial isolates tested, 83 (41.5%) were confirmed as S. aureus of which 51 (61.45%) were oxacillin resistant (ORSA). Out of 51 isolates 26 had single resistance (oxacillin resistance), while 25 had double resistance (oxacillin & methicillin) resistance (MRSA/ORSA).The pathogen city of S. aureus, is related to the production of a wide variety ofexoproteins, including alpha and beta haemolysins which contributes to its ability to cause diseases in many mammalian species [37]. The incidence of haemolytic S.aureus isolated from animal and human samples in the present study was 100℅ of S.aureus isolates were haemolytic on sheep blood agar. It was found that characterization of haemolysinphenotypically based on haemolysis pattern of Staphylococcus aureuson sheep blood agar plate revealed only an alpha haemolysis pattern (18%), beta haemolysis (27%) and gammahaemolysis (54%) [38]. Meticillin resistance is conferred by carriage of the mecA gene [39],which is carried by a mobile exogenous genetic element known as the staphylococcal cassette chromosome mec(SCCmec) [40].In the present study resistance to methicillin was determined by the methicillin disk susceptibility test and confirmed by mecA by PCR [41]. All MRSA isolates in the present either from human or animal origin were mecA gene positive by PCR. While, 80℅ of non-MRSA isolates of human origin were mecA gene positive by PCR. It is interesting to note that 4/5 human non-MRSA isolates were positive for mec A gene by PCR. While, all 5 human MRSA isolates were positive for mec A gene by PCR and the same was found concerning the 5 animal MRSA isolates. The mecA gene which lies in the SCCmec resistance island [42], were found to be carried by 95% of the isolates that display a phenotype of methicillin resistance and was detected in all multiresistant S. aureus isolates which agrees with the results in the present study [43].Habeeb et al. [15] found that PCR data showed that all isolates of MRSA typed in his study were positive for mec A gene where he verified genetic resistance to methicillin by PCR for detection of mecA gene, which was in agreement with the present study results. Regarding the incidence of hlg gene in MRSA and non-MRSA isolates among animal and human samples by PCR were represented in MRSA isolates of human origin with hlg gene PCR positive results by60℅ which is higher than those non-MRSA isolates of human origin which was 40℅. On the other hand, these results showed that the highest percent of hlg gene PCR positive results were represented in MRSA isolates of animal origin (80℅).On comparison between the results of haemolysis on sheep blood agar and the presence of hlg gene among MRSA and non-MRSA isolates. We found that, all strains showed haemolysis on blood agar from human and animal were positive for hlg by PCR. While, those which did not show haemolysis on sheep blood agar, did not possess hlg gene by PCR. Amplification of the gene encoding haemolysin of S. aureus with specific primers showed hla genes with percentage of 81,81 and hla combined with hlb genes in the percentage of 18,18 [38].While, on comparison between the results of Methicillin resistance and the presence of mecA gene among MRSA and non-MRSA isolates. It was clear that human no. 7 (non-MRSA) isolate which showed sensitivity to Oxacilline bacteriologically reveled positive PCR for mec A gene presence. Also, Human no. 9, Human no.11 and Human no.12 (non-MRSA) isolates showed the same variance. On the other hand, the rest 11 isolates out of total 15 isolates gave identical Oxacilline sensitivity PCR result concerning mecA gene presence either by sensitivity or resistance to those results of traditional disc diffusion test. As a result, there is differences between disc diffusion oxacilline resistance results and PCR results for the mec A gene.In contrast to the present study results, mecA gene was present in all isolates recovered by Wielders et al. [43] and which was resistant to four or more antibiotic. Moreover, this multiresistance was displayed by the most prevalent and geographically widespread MRSA types (types І, ІІa, ІІb, and ІІІb), which together represented 99℅ of the mecA population in Europe. Of the European isolates that appeared to be susceptibletomethicillin in the phenotypic test, 10℅ nevertheless contained mec A, and some of these were multiresistant. In contrast, 5℅ of the phenotypically methicillin- resistant isolates did not carry mecA and displayed low-level resistance (>8μg /ml). Rushdy et al. [36] provided an evidence for the presence of mecA gene at 533 bp in all methicillin resistant isolates. This is supported by the work of Murakami et al. [21], Perez-Roth et al. [44] and Japoni et al. [45] who all detected a DNA fragment of 533 bp in all mecA gene in positive methicillin resistant S. aureus, which was absent in susceptible strains, which all disagreed with results of the present study. That’s because the used primers are different from the one used in the present study [21, 44, 45].In the present study, PCR for the detection of mecA gene and hlg gene among MRSA and non-MRSA strains isolated from animal and human samples was performed. There were 5 MRSA isolates positive mecA gene and in the same time negative for hlg gene.In conclusion, virulent drug resistant MRSA was isolated from human and animals. Current data strongly suggest that MRSA can move between people and animals in households, farms and hospitals whether the individuals are colonized or infected. Farther research is needed to understand the frequency of this cross-species transmission and its risk to animal and human populations. Strict hygienic and preventive measures are needed among animals and human populations and during food processing to avoid colonization of MRSA isolates.
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML