| [1] | Dabiza, N., and El-Deib, K., 2007, Biochemical evaluation and microbial quality of Ras cheese supplemented with probiotic strains., Polish J. food and nutrition sciences, 57(3), 255-300. |
| [2] | Hattem, H. E, Taleb, A. T., Manal, A. N., and Hanaa, S. S., 2012, Effect of pasteurization and season on milk composition and ripening of Ras cheese., J. Brewing and Distilling, 3(2), 15-22. |
| [3] | Hayaloglu, A. A., and S., Kirbag, 2007, Microbial quality and presence of moulds in Kuflu cheese., Inter. J. Food Microbiol., 115, 376–380. |
| [4] | Gandomi, H., Misaghi, A., Basti, A. A., Bokaei, S., Khosravi, A., Abbasifar, A., and Javan, A. J., 2009, Effect of Zataria multiflora Boiss. essential oil on growth and aflatoxin formationby Aspergillus flavus in culture media and cheese., Food and chemical toxicology, 47, 2397–2400. |
| [5] | Speare, R, Thomas, A. D., Shea, P. O. and Shipton, W. A., 1994, Mucor amphibiorum in the toad, Bufo marinus, in Australia., Journal of wildlife diseases, 30(3), 399-407. |
| [6] | Fouassier, M, Joly, D., Cambon, M., Lafeuille, H. P. and Condat, P., 1998, Geotrichum capitatum infection in a neutropenic patient. Case report and literature review., Rev. Med. Interne, 19,431-433. |
| [7] | Mori, T, Matsumura, M., Yamada, K., Irie, S., Oshimi, K., Suda, K., Oguri, T., and Ichinoe, M., 1998, Systemic aspergillosis caused by an aflatoxin-producing strain of Aspergillus flavus., Med. Mycol., 36,107-112. |
| [8] | Anstead, G. M., Sutton, D. A., Thompson, E. H., Weitzman, I., Otto, R. A., and Ahuja, S. K., 1999, Disseminated zygomycosis due to Rhizopus schipperae after heatstroke., J Clin Microbiol., 37, 2656-2662. |
| [9] | Kontoyiannis, D. P, Lewis, R. E., May, G. S., Osherov, N., and Rinaldi, M. G. 2002, Aspergillus nidulans is frequently resistant to amphotericin B., Mycoses, 45,406-407. |
| [10] | Steinbach, W. J., and Stevens, D. A., 2003, Review of newer antifungal and immunomodulatory strategies for invasive aspergillosis., Clin. Infect. Dis, 37,157-187. |
| [11] | Balajee, S. A., Lindsley, M. D., Iqbal, N., Ito, J., Pappas, P. G. and Brandt, M. E., 2007, Nonsporulating clinical isolate identified as Petromyces alliaceus (Anamorph Aspergillus alliaceus) by Morphological and Sequence-Based Methods., J. Clin. Microbiol., 45(8), 2701–2703. |
| [12] | Ghibaudo, G., and Peano, A., 2010, Chronic monolateral otomycosis in a dog caused by Aspergillus ochraceus., Veterinary dermatology, 21(5), 522–526. |
| [13] | Oxoid, 2006, The Oxoid Manual. 9th ed., by OXOID Limited, Wade Road, Basingstoke, Hampshire RG24 8PW, England, UK. |
| [14] | Ronald, M. A., 2006, Hand Book of Microbilogical Media for the Examination of Food. CRC Taylor and Francis Group Boca Raton London New York. |
| [15] | APHA, (American Public Health Association), 1998, Standard Methods for the Examination of Water and Wastewater 20th ed., APHA, Inc., New York. |
| [16] | Kure, C. F., and Skaar, I., 2000, Mould growth on the Norwegian semi-hard cheeses Norvegia and Jarlsberg., Inter. J. Food Microbiol., 62, 133–137. |
| [17] | Raper, K. E., and Fennel, D. I., 1965, The genus Aspergillus. Williams and Wilkins, Baltimore. |
| [18] | Harrigan, W. F., and McCance, M. E., 1976, Laboratory Methods in Food and Dairy Microbiology (2nd ed.), Academic Press, New York, NY, USA. |
| [19] | Buchta, V., and M., Otcenasek, 1988, Geotrichum candidum an opportunistic agent of mycotic diseases., Mycoses, 31,363-70. |
| [20] | Yoshida, K., Ando, M., Ito, K., Sakata, T., Arima, K., Araki, S., and Uchida, K., 1990, Hypersensitivity pneumonitis of a mushroom worker due to Aspergillus glaucus., Arch Environ Health, 45,245-7. |
| [21] | Chung, K. K. J., and J. E., Bennett, 1992, Medical mycology, (2nd ed.), Lea and Febeiger, USA. |
| [22] | Pitt, J. I., and A. D., Hocking, 1997, Fungi and Food Spoilage., Blackie Academic and Professional, London, UK. |
| [23] | Frater, J. L., Hall, G. S., and Procop, G. W., 2001, Histologic features of zygomycosis - Emphasis on perineural invasion and fungal morphology., Arch. Pathol. Lab. Med., 125,375-378. |
| [24] | Buommin, N. R, Filippis, E. D., Lopez-Gresa, A., Manzo, M., Carella, E., Petrazzuolo, A., and Tufano, M. M. A., 2009, Bioprospecting for antagonistic Penicillium strains as a resource of new antitumor compounds., World J. Microbiol., 24(2), 185–95. |
| [25] | Florez, A. B., and B., Mayo, 2006, Microbial diversity and succession during the manufacture and ripening of traditional, Spanish, blue-veined Cabrales cheese, as determined by PCR-DGGE., International Journal of Food Microbiology, 110, 165–171. |
| [26] | Santi, M. D., M., Sisti;, E., Barbieri, G., Piccoli, G., Brandi, and V., Stocchi, 2010, A combined morphologic and molecular approach for characterizing fungal microflora from a traditional Italian cheese (Fossa cheese)., Inter. Dairy J., 465-471. |
| [27] | Cathrine, F. K., and I., Skaar, 2000, Mould growth on the Norwegian semi-hard cheeses Norvegia and Jarlsberg., Inter. J. Food Microbiol., 62, 133–137. |
| [28] | Kure, C. F., Y., Wasteson, J., Brendehaug, and I., Skaar, 2001, Mould contaminants on Jarlsberg and Norvegia cheese blocks from four factories., Inter. J. Food Microbiol., 70, 21–27. |
| [29] | Florez, A. B., P. A., Martin, T. M. L., Diaz, and B., Mayo, 2007, Morphotypic and molecular identification of filamentous fungi from Spanish blue-veined Cabrales cheese and typing of Penicillium roqueforti and Geotrichum candidum isolates., International Dairy Journal, 17, 350–357. |
| [30] | Pattono, D., A., Grosso, P. P., Stocco, M., Pazzi, and G., Zeppa, 2013, Survey of the presence of patulin and ochratoxin A in traditional semihard cheeses., Food Control, 33,54-57. |
| [31] | Diaz, L. T. M., R. M. T., Blanco, G. M. C., Arias, G. M. L., Fernandez, and G., Lopez, 1996, Mycotoxins in two Spanish cheese varieties., Inter. J. Food Microbiol., 30, 391-395. |
| [32] | Erdogan, A., M., Gurses, and S., Sert, 2003, Isolation of moulds capable of producing mycotoxins from blue mouldy Tulum cheeses produced in Turkey., Inter. J. Food Microbiol., 85, 83-85. |
| [33] | Asta, D. C., D. J., Linder, G., Galaverna, N. E. A., Dossena, and R., Marchelli, 2008, The occurrence of ochratoxin A in blue cheese., Food Chemistry, 106, 729-734. |
| [34] | Abdel-All, S. M, M. A., Abd-El-Ghany, and M. M., Motawee, 2008, Inhibition of Aspergillus growth and aflatoxins production in some dairy products. The 3ed annual conference of quality education development in Egypt and the Arab region to achievement the requirements of job markets in the globalization age (strategic vision). Faculty of Specific Education Mansoura University, 1108-1120. |
| [35] | Berlkten, D., and M., Kivanc, 2012, Fungal contamination of some food, their mycotoxin and effects of antifungal agents on these fungi., Microbes in Applied Research, Current advanced and challenges, World Scientific Publishing Co. |
| [36] | Nasser, L. A., 2001, Fungal contamination of white cheese at the stage of consumption in Saudi Arabia., Pakistan J. Biological Sciences, 4(6), 733-735. |
| [37] | Ando, H., K., Hatanaka, I., Ohata, Y. Y., Kitaguchi, A., Kurata, and N., Kishimoto, 2012, Antifungal activities of volatile substances generated by yeast isolated from Iranian commercial cheese., Food Control, 26,472-478. |
| [38] | Sharma, J., A., Singh, R., Kumar, and A., Mittal, 2013, Partial purification of an alkaline protease from a new strain of Aspergillus oryzae AWT 20 and its enhanced stabilization in entrapped Ca-alginate beads., Internet J. Microbiol., 2(2), 1-14. |
| [39] | Hassan, H. N., and S. A., El-Deeb, 1988, The inhibitory effect of water and acetone extraction of certain plants as anti-fungal agents on growth and aflatoxins production by molds and yeast isolated from cheese ripening rooms., J. Agric. Res., Tanta Univ., 14, 162-171. |
| [40] | El-Deeb, S. A., E. E., Kheadr, N., Zaki, and Y. M., Shoukry, 1992, Formation and the penetration of aflatoxin in experimental cheese by Aspergillus parasiticus., Egyptian J. Food Sci., 15-20. |
| [41] | Anwar, A., and Y., Sabah, 2010, The components effect of cinnamon oil extracted in inhibition of isolated molds from Kashkawan cheese., Damascus J. of agricultural Science, 26(2), 287-300. |
| [42] | Barrios, M. J., L. M., Medina;, M. G., Cordoba, and R., Jordano, 1997, Aflatoxin producing strains of Aspergillus flavus isolated from cheese., J. Food Protection, 192-194. |
| [43] | Taniwaki, M. H., A. D., Hocking, J. I., Pitt, and G. H., Fleet, 2001, Growth of fungi and mycotoxin production on cheese undermodified atmospheres., Inter. J. Food Microbiol., 68, 125–133. |
| [44] | Cheong, E. Y. L., A., Sandhu, J., Jayabalan, T. T. K., Le, N. T., Nhiep, H. T. M., Ho, J., Zwielehner, N., Bansal, and M. S., Turner, 2014, Isolation of lactic acid bacteria with antifungal activity against the common cheese spoilage mould Penicillium commune and their potential as biopreservatives in cheese., Food Control, 46, 91–97. |
| [45] | Lynch, K. M., A. M., Pawlowska, B., Brosnan, A., Coffey, E., Zannini, A., Furey, and P. L. H., McSweeney, D. M., Waters, and E. K., Arendt, 2014, Application of Lactobacillus amylovorus as an antifungal adjunctto extend the shelf-life of Cheddar cheese., Inter. Dairy J., 34,167-173. |


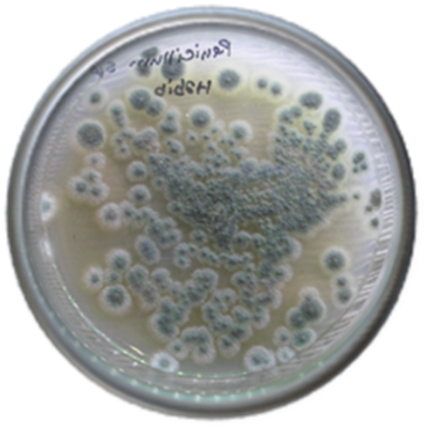
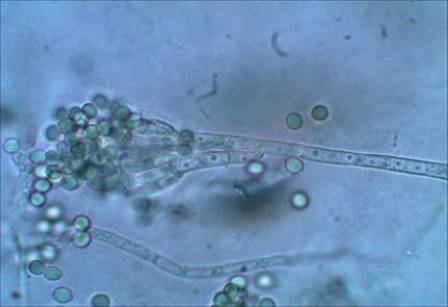
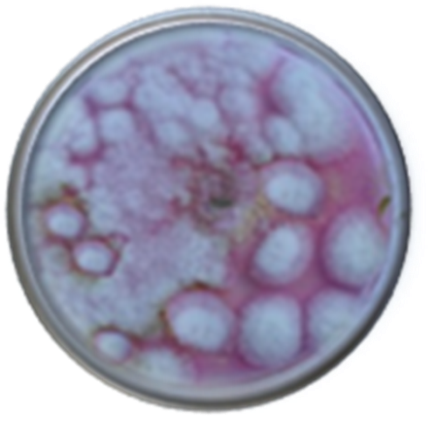

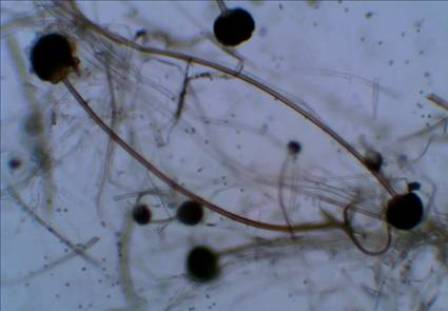
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML