-
Paper Information
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
Journal of Microbiology Research
p-ISSN: 2166-5885 e-ISSN: 2166-5931
2014; 4(2): 104-111
doi:10.5923/j.microbiology.20140402.10

Evaluation of Diagnostic Techniques for the Detection of Brucella spp in Camel Sera and In-Vitro Testing Their Susceptibility against Antimicrobial Agents
1Department of Bacteriology, Mycology and Immunology, Faculty of Veterinary Medicine, Sadat City University, Egypt
2Department of Public Health (Microbiology), College of Public Health and Health Informatics, Qassim University, Saudi Arabia
Correspondence to: Ayman El Behiry , Department of Bacteriology, Mycology and Immunology, Faculty of Veterinary Medicine, Sadat City University, Egypt.
| Email: | 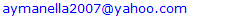 |
Copyright © 2014 Scientific & Academic Publishing. All Rights Reserved.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/

Brucellosis is a zoonotic disease and represents one of the most common public health problems worldwide. For diagnosis and treatment of the disease, sensitive and accurate methods are required. Although serological techniques are considered the main diagnostic method of camel brucellosis, these tests have been immediately transferred from cattle without sufficient legalization. Until now, scanty data on utilization of real-time PCR for identification of Brucella in camel is available. Therefore, this study was conducted to evaluate the serological techniques and real-time PCR for the detection of Brucella species and to determine in vitro antimicrobial resistance of these strains to frequently used anti-Brucella agents. A total of 410 serum samples were identified genotypically by real-time PCR and serologically by Rose Bengal Plate Test (RBPT), Slow Agglutination Test (SAT), Complement Fixation Test (CFT), competitive enzyme-linked immunosorbent assays (cELISA) and fluorescence polarization assay (FPA). In vitro activities of various antimicrobials were also evaluated by the E test method. According to the serological results, FPA demonstrated the maximum number of positive cases (84.87%) followed by CFT (79.75%), SAT (77.56%), RBPT (76.58%) and cELISA (75.36%). However, 88.53% of all cases were identified by real-time PCR. The lowest MIC90 values were observed in trimethoprim-sulfamethoxazole (0.195 µg/ml), doxycycline (0.390 µg/ml), chloramphenicol (0.781 µg/ml) and amikacin (1.562 µg/ml). In contrast, amoxicillin/clavulanic acid, streptomycin, rifampin, ceftazidime, cefotriaxone and ceftriaxone had high MIC50 and MIC90 against most of identified Brucella strains. Based on the results of this study, it is recommended to use a real-time PCR technique in combination with at least one serological test for precise diagnosis of camel brucellosis in developed and less-developed countries. In addition, using a combination of rifampin, doxycycline with trimethoprim-sulfamethoxazole is considered promising anti-Brucella agents. Camels must be included in nationwide plans for eradication and control of brucellosis in endemic regions.
Keywords: Brucella spp., Real-time PCR, Antimicrobial resistance, Camels, Camel ranchers
Cite this paper: Ayman El Behiry , Evaluation of Diagnostic Techniques for the Detection of Brucella spp in Camel Sera and In-Vitro Testing Their Susceptibility against Antimicrobial Agents, Journal of Microbiology Research, Vol. 4 No. 2, 2014, pp. 104-111. doi: 10.5923/j.microbiology.20140402.10.
Article Outline
1. Introduction
- Brucellosis is a prevalent zoonotic disease affecting both humans and animals [1, 2]. It is found to be one of the most common public health problems all over the world [3]. Gram-negative bacteria coccobacilli of the genus Brucella is considered the main cause of this disease in man and animals. The most common types of Brucella species concentrated in the Near East region are Brucella melitensis and Brucella abortus [4]. Different countries such as Saudi Arabia, Kuwait, Oman, United Arab Emirates, Iraq, Iran, Sudan, Egypt, Libya and Somalia recorded high incidence of brucellosis in camels [5]. Consumption of milk and meat from infected camels with Brucellosis led to a high frequency of human Brucellosis cases and consequently severe public health problem has established [6]. Precise diagnosis of livestock and humans brucellosis is considered the keystone for its correct abolition and manages [7]. In general, the diagnosis of brucellosis is somewhat difficult as the disease may have an incubation period varying from 5 days to 5 months and can progress in various forms: acute, chronic or asymptomatic [5, 8]. Although serological techniques of brucellosis have been used for detection of brucellosis in camels, they are neither sufficiently sensitive nor specific as a result of an inadequate immune status of the host [5]. Isolation of the causative agent is considered the main particular diagnostic technique; however this method is prolonged and low sensitive, particularly in the chronic phase of the infection [7]. The progress of accurate diagnostic techniques for direct recognition of Brucella species is increasingly drawing attention to reduce the problems induced by conventional methods. Diagnosis of brucellosis based on the standard techniques such as culture and phenotypic characterization, is hard, prolonged, increase the possibility of infection, and may make a negative results. Consequently, serological assays are frequently used for diagnosis of animal brucellosis particularly in cattle, sheep, goats and camels, but cross-reactions with other gram-negative bacteria till now represent a big problem (9). Rose Bengal Plate Test (RBPT), Complement Fixation Test (CFT), and Slow Agglutination Test (SAT) are frequently utilized for the detection of antibodies specific to Brucella spp [4, 5, 10]. The susceptibility of RBPT completes the requirement for observation of free areas at flock level, nevertheless it is believed that merely the combination of RBPT and CFT in infected herds with brucellosis be capable of obtain accurate individual sensitivity in test-and-slaughter programs. Additionally, World Organization for Animal Health (OIE) suggested that CFT is a test approved all over the world [11]. This test is considered as a high-quality test when correctly used, however, it has lots of practical drawbacks such as time consuming and difficult to standardize [5, 8, 9]. The above revealed techniques can’t distinguish between antibodies formed subsequent vaccination and those results from infection [12]. Competitive Enzyme-linked immunosorbent assays (cELISA) have been applied to solve these troubles. Moreover, ELISA could detect Brucella carriers, which were sero-negative by RBT, SAT and CFT [5, 8, 13]. Even though SAT is considered the gold standard technique in the serological diagnosis of brucellosis, cELISA technique is frequently utilized in the clinical laboratories, particularly in non endemic areas. Commercial ELISA kits reliably detect anti- Brucella antibodies and the results are reliable with the SAT and CFT [9]. Currently, the fluorescence polarization assay (FPA) is a serological method applied for diagnosis of brucellosis both in humans and animals due to it is a quick, homogenous, species-independent technique, which was initially progressed and authenticated for the recognition of antibodies specific to Brucella abortus in cattle. FPA has numerous realistic benefits over the other techniques. Presently, it has to become the most important technique within the routine testing methods of most National Brucellosis Reference Laboratories. The requirements of FPA need minimal preparations and can be done in few minutes [11, 14]. It is suggested that the different serological techniques utilized for Brucellosis in animals, especially cattle may also be adequate for diagnosis of Brucellosis in camels. Nevertheless, no legalization for camel sera was carried out yet. Although there are numerous researches on detection of DNA extracted from Brucella spp by PCR from pure culture, only a few researches have been carried out in camels with clinical or field samples [5]. The advantages of PCR are numerous such as it is more specific than blood cultures and more accurate than serological assays. Navarro et al. [15] and Al Dahouk et al. [16] found that a diversity of PCR assays targeting various gene loci have been successfully utilized for the molecular diagnosis of Brucellosis. The sensitivity of diagnosis can be improved by using real-time PCR techniques, which has the ability to detect as few as five pathogens per reaction [15, 17]. Moreover, real-time PCR helps molecular diagnosis of clinical specimens and giving results within a few hours. In fact, Gwida et al. [5] indicated that there is a little information about the application of real-time PCR for identification of the different isolates of Brucella isolated from the blood serum samples of camels. As, the Brucella spp. is an intracellular gram negative bacteria that infects host phagocytic cells as a result, specialized antibacterial agents that are able to perforate the phagocytic cells and work inside their cytoplasm are required for the treatment of animal and human brucellosis [18, 19]. Accordingly, a limited number of antimicrobial agents are efficient against this type of bacteria. Acute Brucellosis may relapse or become chronic disease due to lack of a sufficient long-term antimicrobial treatment, [8, 20, 21]. The World Health Organization (WHO) suggested that the most excellent mixture antibiotics used for human brucellosis are doxycycline along with either rifampin or streptomycin [22]. Although Brucella isolates are frequently believed susceptible to these antimicrobial agents, individual cases of antibiotic resistance and recurrence of infection have been reported. Nevertheless, routine antimicrobial susceptibility testing is habitually not carried out for Brucella as a result of its difficult growth necessities; danger of laboratory acquired infections and requires for biological safety level 2or 3 precautions [22]. In the current study, the primary objective was to detect Brucella antibodies in camel sera using different sociological techniques. The second objective was to determine their resistance to various antimicrobial agents using E-tests.
2. Material and Methods
2.1. Sample Collection
- A total of 321 blood samples were collected from diseased and apparently health camels (camelus dromedarius) at Al-Qassim regions which lie in the middle area of the Kingdom of Saudi Arabia (KSA). Moreover, human blood samples were obtained from eighty nine selected males from the peoples in the area neighboring the camel farms, who worked and/or had a direct contact with the camels under study.
2.2. Isolation and Identification of Brucella spp
- 5 ml of peripheral blood were taken from each animal and human and divided into two parts. One part was collected in EDTA and the serum was separated from the second part and stored at –20 oC until processing with serological tests. The first part of the blood with anticoagulant was inoculated into modified Farrell’s serum dextrose agar which used to isolate Brucella spp. according to standard measures [7]. Modified Farrell’s serum dextrose agar with 5% horse serum, 1% dextrose, and the following antibiotics (added to 1 L medium): cycloheximide (100 mg), bacitracin (25,000 IU), polymyxin B sulfate (5,000 IU), vancomycin (20 mg), nalidixic acid (5 mg), and nystatin (100,000 IU), was used for primary isolation of brucellae. In the presence of 5–10 % carbon dioxide, plates were inoculated with the sample and incubated aerobically at 37 °C. These plates were examined 3–7 days after inoculation for bacterial growth. The suspected colonies were subcultured again for purity on serum dextrose agar. Based on the standard measures obtained by Alton et al. [7], identification of these isolates was carried out. Primarily, all isolates were checked for Gram and modified Ziehl–Neelsen (MZN) staining. Subsequent biochemical tests for oxidase, catalase, urease production, hydrogen sulphide production, carbon dioxide requirement, growth on media containing basic fuchsin and thionin (20 μg/ml), and agglutination by monospecific antisera (A, M, R; Anses, Paris) was carried out.
2.3. Serological Tests
- All serum samples of camels and camel ranchers were examined by RBPT, SAT, CFT, cELISA and FPA. Antigens utilized for RBPT, SAT, and CFT were purchased from BIO-RAD Company, Marnes-la-Coquette, France. Positive and negative control sera are the national reference sera standardized according to the World Organization for Animal Health (OIE). According to the Manual of Standards for Diagnostic Tests and Vaccines [23], RBPT was performed using antigen purchased from BIO-RAD Company, Marnes-la-Coquette, France. SAT was carried out in microtitre plates [7]. Samples are screened more than 30 I.U. Per milliliter was believed to be positive. The reagents utilized in the CFT were standardized and the test was carried out according to the guidelines recommended by OIE [23]. Any serum showing a value ≥ 20 ICFTU per millilitre was believed to be positive. The cELISA was carried out and results were calculated according to the instructions of the manufactures using Svanovir™ Brucella-Ab ELISA kit purchased from the company, Svanovia Biotech AB Uppsala, Sweden. Finally, FPA was carried out and the results were recorded according to the instructions of the manufacturer (Diachemix, LLC, 223 North Water Street, Suite 500 Milwaukee, WI 5320-25707, U.S.A.). In brief, the protocols for these techniques used in camel and human brucellosis were the same as utilized for bovine brucellosis.
2.4. DNA Extraction
- DNA was purified using the High Pure PCR Template Preparation Kit (Roche Diagnostics GmbH, Sandhofer Street 116, DE-68305 Mannheim, Germany) according to manufacturer’s instructions. The degree of purity and concentrations of extracting DNA were detected by the Nano-Drop ND-2000 UV–Vis Spectrophotometer (Thermo Fisher Scientific, NanoDrop products Wilmington, DE 19810, USA), and preparations were stored at −20°C for later investigation.
2.5. Real-time PCR
- Real-time PCR for the genus specific Brucella was carried out on DNA extracted from serum samples of camels and camel handlers using the following primers (5`GCTCGGTTGCCAATATCAATGC3`) as forward primer and (5` GGGTAAAGCGTCGCCAGAAG 3`) as the reverse primer together with genus specific probe (5`AAATCTTCCACCTTGCCCTTGCCATCA 3`) [24]. The primers and probes were designed and purchased from Qiagen, Germany. The REAL-TIME PCR technique was carried out by the use of the TaqManTM Universal Master Mix (Qiagen GmbH, Hilden, Germany) according to the manufacturer guidelines using a 12.5 µl master mix, 0.75 μl of each primer and 0.25 μl TaqMan probe. 2 μl of genomic DNA was utilized as target and nuclease-free water (Qiagen GmbH, Hilden, Germany) was added up to a total reaction volume of 25 μl. The REAL-TIME PCR reaction was performed in duplicate in microtiter plates with 96-well using a thermocycler system with the following run setting: 1 cycle of 50°C for 2 min., 1 cycle of 95°C for 10 min., followed by 50 cycles of 95°C for 25s and 57°C for 1 min.
2.6. Antimicrobial Resistance Testing
- A total of 200 strains of Brucella species, both of human and animal origin were included in the current study. In vitro susceptibility of sixteen antimicrobial agents against Brucella strains was carried out according to the Minimal Inhibitory Concentration (MIC) values. For this reason, various techniques such as microbroth dilution, agar dilution and E-test can be used. In the present study, the Epsilometer test (E-test) (BioMérieux, 69280 Marcy l’Etoile, France) method was used due to it is an accurate, rapid, less labor-intensive and more practical method [19]. Susceptibility to Tetracycline, Doxycycline, Ciprofloxacin, Streptomycin, Rifampin, Gentamycin,Trimethoprim-sulfamethoxazole, Chloramphenicol, Ceftriaxone, Cefotaxime, Ceftazidime, Cefepime, Amoxicillin/clavulanic acid, Ofloxacin, Ciprofloxacin and Amikacin were evaluated. Suspension of the growing bacterial colonies in Mueller-Hinton broth (Oxoid®, Hampshire, UK) with a turbidity equivalent to that of a 0.5 McFarland standard was inoculated at 37°C for two days by gently sloping a sterile swab over 5% sheep blood-enriched Mueller-Hinton agar (Oxoid®, Hampshire, UK) plates. Detection of the MIC was carried out according to strategy of the Clinical Laboratory Standards Institute’s (CLSI) for Brucella and slowly growing bacteria species. Briefly, Mueller- Hinton agar plates supplemented with 5% sheep blood were inoculated with bacterial suspensions calibrated to 0.5 McFarland standard turbidity and E-test strips were applied. The plates were placed in a 5% CO2 incubator for 48 hours and the resulting growth was examined to determine the MIC for the above mentioned antibiotics. Three Brucella reference strains (Brucella abortus 544, Brucella melitensis 16M, and Brucella suis 1330) were utilized as controls for serological and REAL-TIME PCR assays as well as antimicrobial susceptibility testing. In addition, these Brucella reference strains, other strains such as Staphylococcus aureus ATCC 29213 and Escherichia coli ATCC 25922 will be also utilized as a control strain for antimicrobial susceptibility testing.
3. Results
3.1. Serological and Real-time PCR Tests
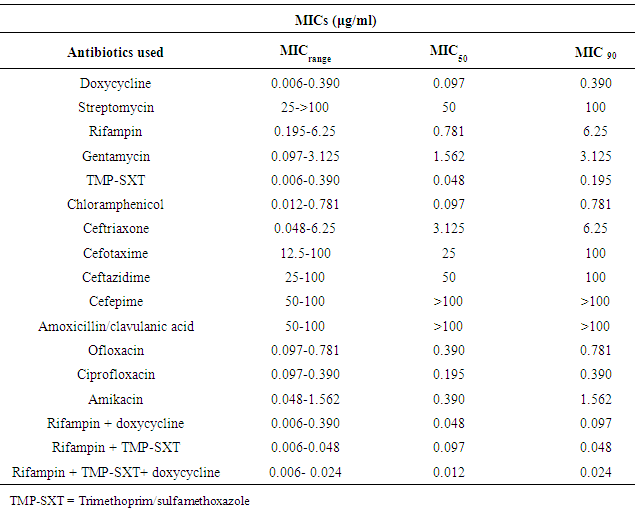
- As can be seen from table 1, the REAL-TIME PCR was able to detect the gene encoding of bcsp3 for both dromedary camels (diseased and apparently healthier) and camel ranchers in 88.53% (363/410) samples. After examination of all samples by FPA, the positive samples were 84.87% (348/410), while 79.75% (327/410), 77.56% (318/410), 76.58% (314/410) and 75.36 (309/410) of the camel and human samples were positive to CFT, SAT, RBT and cELISA tests, respectively. From the previous results we found that 15 samples were positive to REAL-TIME PCR and negative in all serological tests. The conformity between the results achieved by FPA and that for bcsp31 real-time PCR is demonstrated in (Table 1). FPA was positive in 348 samples out of 410 (84.87%) that were also positive by bcsp31 real-time PCR. As confirmed by bcsp31 real-time PCR, the presence of DNA of Brucella or anti-Brucella antibodies verified by two various serological assays were believed as an evidence for a possible danger of customers when consuming products of these animals. The board of sera which perform no less than one of these criteria was thought to be the “gold standard”. Based on this description (positive by real-time PCR or two various serological assays) 309 samples had to be represented as positive. For both human and animal samples, 363 samples out of 410 were identified by real-time PCR with a sensitivity of 88.53%. This means real-time PCR was more accurate than all serological tests when used in the detection of different Brucella species.
|
3.2. Antimicrobial Resistance of Brucella Species
- A total of 200 isolates of Brucella species were tested against different antimicrobial agents using the E test method. According to the results, the range of the Minimum Inhibitory Concentration (MICrange), Minimum Inhibitory Concentration required to inhibit the growth of 50% (MIC50) and 90% (MIC90) Brucella isolates were demonstrated in table 2. Nearly all isolates of Brucella species were resistant to amoxicillin/clavulanic acid, streptomycin, ceftazidime, cefotaxime and ceftriaxone. The MICrange values of doxycycline (0.006-0.390 μg/ml),trimethoprim-sulfamethoxazole (0.006-0.390 μg/ml), chloramphenicol (0.012-0.781 μg/ml), ciprofloxacin (0.097-0.390 μg/ml), ofloxacin (0.097-0.781 μg/ml) and amikacin (0.048-1.562 μg/ml) were low against all isolates of Brucella species. The MICrange values of a combination of rifampin with doxycycline (0.006-0.390 μg/ml); rifampin withtrimethoprim-sulfamethoxazole (0.006-0.048 μg/ml) or rifampin, doxycline with trimethoprim-sulfamethoxazole (0.006-0.024 μg/ml) were lower than using of these antimicrobial agents alone (Table 2). Whereas, MIC values of rifampin, gentamycin and ceftriaxone were moderate against all isolates. From the previous results, we concluded that the isolates of Brucella isolated from both camels and camel ranchers were highly sensitive to doxycycline, trimethoprim-sulfamethoxazole and chloramphenicol. In contrast, nearly all isolates were highly resistant to streptomycin, amoxicillin/clavulanic acid.
|
4. Discussion
- Control of animal and human brucellosis depends on the perfect techniques used for detection, identification and treatment of the causative agent. Nevertheless, the diagnosis of brucellosis in camels is still frequently difficult. Characteristic clinical symptoms of animal brucellosis especially in camels are often absent and till now the diagnostic methods are not assessed. In the current research, all camels at the time of samples taking were apparently normal and according to the owner’s data, none had formerly revealed clinical signs of brucellosis. These facts point to that many infected camels might be carriers for brucellosis without the appearance of clinical signs and their products may lead to a severe public health problem for consumers. Moreover, in Saudi Arabia human brucellosis have increased attention in camel brucellosis and encouraged camel breeder to bring samples for examination. In the current study, sporadic cases of abortion in pregnant camels and Mediterranean fever in some camel-keeping families were recorded in Al-Qassim region. The occurrence of abortion in camels on the Al-Qassim farms was about 10%, in the lack of any other symptoms. Furthermore, approximately 20% of the camel ranchers and milkers in the Al-Qassim farms had Mediterranean fever as a result of infection with Brucella melitensis at the time of the current investigation.
4.1. Diagnosis of Brucellosis by Serological Assays
- Various serological tests were applied in the current study to detect the most suitable technique used for screening the infection and identifying its cause in animals and human beings. 410 blood serum samples were collected arbitrarily from camels (321 samples) and camel ranchers (89 samples) in Al-Qassim region. The obtained results indicated that 84.87%, 79.75%, 77.56%, 76.58% and 75.36% of the camel and human sera examined were positive by FPA, CFT, SAT, RBT and cELISA, respectively. From these results, the current study indicated that FPA and CFT are better than the other tests. This result is in agreement with that formerly reported [5, 25, 26]. These authors confirmed that the FPA and CFT are the most broadly applied tests for animal and human brucellosis. Therefore, in the current study, it is recommended to utilize cELISA technique for observation and monitoring of brucellosis both in humans and animals especially camels. In addition, using of FPT and CFT are very important in confirmation of positive cases. Nevertheless, molecular tests and techniques are also required to be assessed. This applies particularly to utilize of reverse transcription real-time PCR for identification of an active infection.
4.2. Molecular Diagnosis by Real-time PCR
- Fast, ultimate and precise diagnosis of human and animal brucellosis is extremely significant in a correct way of elimination protocols [27, 28]. Currently, real-time PCR techniques are simple to be achieved, highly sensitive, and give more specificity for identification of different types of microorganisms. PCR and real-time PCR are considered as alternative methods for the failure of culturing and identification of Brucella spp. by traditional methods [5, 29]. As can be seen from table 1, 87.53% of all animal samples (281 animal out of 321) was positive to REAL-TIME PCRs targeting bcsp31 and 92.13% of human samples was positive to real-time PCRs targeting bcsp31 (82 out of 89). According to these results, molecular methods used for identification of Brucella spp. Isolated from both humans and animals might avoid the disadvantages produced by different serological techniques. Therefore, PCR technique has been revealed to be an important technique for identifying DNA of bacteria and affords a promising alternative method for diagnosis of animal and human brucellosis. There are numerous advantages for using real-time PCR over the conventional methods, because it is more rapid and accurate. As well as, it decreases the danger of spreading the disease to people working in the laboratory. Whereas PCR directly identifies the DNA of the bacteria, serological assays are depending upon the increasing and/or decreasing titers of antibodies throughout the various clinical stages of brucellosis. Therefore, real-time PCR is the assay to verify the identification of animals and humans brucellosis especially in less-developed countries. Al -Dahouk et al. [30], Bounaadja et al. [31] and Al-Garadi et al. [32] suggested that real-time PCR is considered a confirmatory technique for brucellosis. High specificity technique is required to decrease the economical losses inherent in the management and the abolition of Brucella infection in camel farms. No proof was provided by the current study to support the utilize of specific serological tests to diagnose Brucella infection because of the low accuracy of the existing serological assays to identify brucellosis in the early phase of the disease when diagnoses depends mostly on detection of either immunoglobulin M (IgM) or immunoglobulin G (IgG) by antigens purchased formerly. The utilization of more than one assay is highly recommended for diagnosis of both human and animal brucellosis [5, 33]. Therefore, using of serological assay and substantiation by a molecular diagnosis, particularly real-time PCR is the most excellent method to manage and eliminate the Brucella infection on camel farms and correct diagnosis of individuals infected with brucellosis. In contrast, one single test is not enough to verify the diagnosis of brucellosis and a combination of two tests should be carried out, preferably the FPT and real-time PCR.
4.3. Antimicrobial Resistance of of Brucella Species
- Treatment of animal and human brucellosis requires antimicrobial agents that can perforate macrophages and can work in the acidic intracellular environment. Isolates resistant to the majority of antibiotics may emerge and lead to treatment failure [34, 35]. The isolates included in this study were collected from various localities of Al-Qassim region. According to the results obtained, nearly all isolates were most resistant to nearly all isolates of Brucella species were resistant to amoxicillin/clavulanic acid, streptomycin, ceftazidime, cefotaxime and ceftriaxone used in this work. Similar results were obtained by Safi & Al-Mariri [35]. In contrast, these results were partially not in agreement with that recorded by [36], where they confirmed that cefotaxime was considered one of the most effective antibiotics used, with MICs ranging from 0.25 to 2 μg/ml. In the current study, the MICrange values of doxycycline, trimethoprim-sulfamethoxazole (0.006-0.390 μg/ml), chloramphenicol (0.012-0.781 μg/ml), ciprofloxacin (0.097-0.390 μg/ml), ofloxacin (0.097-0.781 μg/ml) and amikacin (0.048-1.562 μg/ml) were low against all isolates of Brucella species. a combination of rifampin with doxycycline (0.006-0.390 μg/ml); rifampin with trimethoprim-sulfamethoxazole (0.006-0.048 μg/ml) or rifampin, doxycline with trimethoprim-sulfamethoxazole (0.006-0.024 μg/ml) were lower than using of these antimicrobial agents alone Whereas, MICrange values of rifampin, gentamycin and ceftriaxone had a moderate activity against all isolates. Therefore, doxycycline in combination with rifampin andtrimethoprim-sulfamethoxaz-ole were considered the most effective anti-Brucella agents used. Doxycycline is considered as a gold standard antibiotic by the World Health Organization (WHO) and it became the most frequently approved tetracycline derivative in the treatment of Brucella infections because of its superior pharmacokinetics profile [37]. In several researches, doxycycline was considered to be one of the most effective antimicrobial agent against Brucella strains isolated from both humans and animals [19, 37]. In addition, the current study demonstrated that doxycycline was found to be the most effective agent with the lowest MIC50 and MIC90 values. Trimethoprim-sulfamethoxazole is an additional antimicrobial agent recommended for the treatment of human and animal brucellosis [37]. Earlier studies confirmed that trimethoprim-sulfamethoxazole is considered an effective antimicrobial agent with low MIC levels [34]. Aliskan et al. [38] indicated that trimethoprim-sulfamethoxazole is considered an effective antibiotic with the lowest MIC50 and MIC90 values. In the current research, trimethoprim- sulfamethoxazole was found to be one of the mainly useful antimicrobial agents after doxycycline based on the MIC50 and the MIC90 values. Furthermore, there are two antimicrobial agents named ciprofloxacin and ofloxacin (quinolones group) were used in the present study with low MIC50 and the MIC90 values have a good effect against brucellosis. Fluoroquinolones easily penetrate into the cells and they are efficient against Brucella spp. [15, 33]. Prior trial verified that fluoroquinolones are considered one of the most effective groups of antibiotics against Brucella spp. [34, 37, 39].
5. Conclusions
- Based on the results obtained from the current study, the utilization of real-time PCR technique in combination with at least one serological assay, especially FPA is recommended for accurate diagnosis of camel brucellosis in developed and less-developed countries. Using a combination of two or three antimicrobial agents such as rifampin, doxycycline with trimethoprim-sulfamethoxazole is considered a promising anti-Brucella agent. Furthermore, antibiotic susceptibility patterns of Brucella spp. seams to be fluctuated geographically, hence regional periodic evaluation of vulnerability of isolates to antimicrobial agents has to be suggested. From the above mentioned data camels should be included in nationwide plans for eradication and control of brucellosis in endemic regions.
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML