-
Paper Information
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
Journal of Microbiology Research
p-ISSN: 2166-5885 e-ISSN: 2166-5931
2013; 3(4): 135-142
doi:10.5923/j.microbiology.20130304.02
Variation of Protease Production by the Bacteria (Bacillus fastidiosus) and the Fungus (Aspergillus funiculosus)
Zinnat Shahina1, Mohammad Towhid Hossain2, Mohammad Abdul Hakim2
1Department of Microbiology, University of Science and Technology Chittagong, Bangladesh
2Department of Microbiology, Chittagong University, Bangladesh
Correspondence to: Zinnat Shahina, Department of Microbiology, University of Science and Technology Chittagong, Bangladesh.
| Email: |  |
Copyright © 2012 Scientific & Academic Publishing. All Rights Reserved.
Protease enzymes perform the hydrolysis of proteins or large polypeptide chains into smaller peptides and amino acids, and thus facilitate their absorption by cells which is produced by different types of microorganisms such as bacteria, fungi, yeast and actinomycetes. Commercially they are very important in different industrial levels and products. The aim of this study was to isolate the protease producing bacteria and fungi and to optimize the media composition that supports the maximum protease production. In the present study the variation of protease production by the bacteria (Bacillus fastidiosus, Z8) and the fungi (Aspergillus funiculosus, Za) was observed by determining their optimum conditions e.g. incubation period, temperature, pH, substrate specificity and substrate concentration etc, where the results showed that the fungal isolate Za under study are good producers of protease than the bacterial isolate Z8 though it took longer incubation time (5 days) for crude enzyme production in the presence of fructose as carbon source at temperature 35°C with pH 6.0.This might be an indication that protease production can be variable amongst microbes to microbes.
Keywords: Aspergillus Funiculosus, Bacillus Fastidiosus, Protease, Proteolytic Bacteria, Proteolytic Fungi
Cite this paper: Zinnat Shahina, Mohammad Towhid Hossain, Mohammad Abdul Hakim, Variation of Protease Production by the Bacteria (Bacillus fastidiosus) and the Fungus (Aspergillus funiculosus), Journal of Microbiology Research, Vol. 3 No. 4, 2013, pp. 135-142. doi: 10.5923/j.microbiology.20130304.02.
Article Outline
1. Introduction
- The protease enzymes perform the hydrolysis of large polypeptide chains into smaller peptides and amino acids, and thus facilitate their absorption by cells. Commercially they are very important in different industrial levels and products. Generally these enzymes are isolated from various living sources such as plants, animals, and microbes[1]. A variety of microorganisms such as bacteria, fungi, yeast and actinomycetes are known to produce these enzymes[2]. These proteases have wide applications in leather, laundry, food and waste processing industries[3]. Besides, they are also used in pharmaceuticals, medical diagnostics, lens cleansing, decomposition of gelatin on X- ray films and in textiles as a non-hazardous bioalternative[4-7]. Protease enzymes that are isolated from microbial sources are preferred than the enzymes from plant or animal sources since they possess almost all the characteristics desired for their biotechnological applications[8]. Among the various sources, bacterial proteases are the most significant, compared with animal and fungal proteases. Among bacteria, Bacillus species are considered as specific producers of extracellular proteases[9]. But the main draw back in the production of bacterial proteases is the requirement of cost intensive procedures for the separation of enzymes from cells. On the other hand, enzymes from fungal origin offer an advantage of separation of mycelium by simple filtration. Besides these, fungus can be grown in inexpensive substrates[10]. Fungi elaborate a wider variety of enzymes than bacteria do[11]. The application in the leather industry for dehairing and bating of hides to substitute currently used toxic chemicals is a relatively new development and has conferred added biotechnological importance[12]. The activity of an enzyme is due to its catalytic nature. The enzyme activity depends upon the substrate concentration, pH, temperature and other physico-chemical factors[13]. The present investigation is aimed at optimizing the growth conditions of different bacteria and fungi isolated from different sources to enhance the protease production and to compare the amount of protease produced by them.
2. Materials and Methods
2.1. Microorganisms
- For the isolation of protease producing microbes different sources such as rotten shrimp and spoiled pulse seeds were used.
2.2. Screening of the Isolates
- Primary screening was done by using enrichment technique followed by three methods – Boiled egg albumin degradation, Skimmed milk casein hydrolysis and Gelatin hydrolysis. After primary selection, the organisms were used for secondary screening, where the isolates were examined for the protease activity in liquid medium such as (i) Peptone 2%, yeast extract 1%, dextrose 2%[14], (ii) Tryptone 1%, yeast extract 0.5%, dextrose 0.1%[15], (iii) Gelatin 1%, yeast extract 0.2%, glucose 1%, K2HPO4 0.3% KH2PO4 0.1%, MgSO4.7H2O trace[16] were used.
2.3. Measurement of Enzyme Activity
- Protease activity of selected isolates was done by using the modified method of Hayashi et al[17], as followed by Meyers and Ahearn[18]. In this method crude enzyme solution (3 ml) was incubated with 3ml of citrate phosphate buffer and 3ml of 1% casein at 35°C in a water bath. The reaction was stopped by adding 5ml of 20% TCA. After one hour, the solution was filtered by Whatman no. 40 filter paper (Ashless). One ml enzyme substrate mixture was taken into a test tube and 2ml of 20 % Na2C03 and 1 ml of Folin Ciocalteu Reagent was added and shaken well and waited for 30 minutes. After this fixed time period 6 ml distilled water was added to it and absorbance of the solution was measured at 650 nm in a spectrophotometer. Enzyme activity was calculated by measuring mg of tyrosine equivalent released and compared with the standard. One unit (U) of enzyme activity represents the amount of enzyme required to liberate 1 μg of tyrosine/ml under standard assay conditions.
2.4. Biomass Yield
- For fungal isolate the biomass residue was dried at 70°C to a constant weight and the amount of biomass was calculated by subtracting the weight of filter paper as the supernatant was collected by what man no. 1 filter paper. The yield was expressed as mg g-1 of protein. On the other hand bacterial biomass was determined by measuring the absorbance at 600 nm[19].
2.5. Optimization of Culture Conditions
2.5.1. Effect of Incubation Time, Temperature and Medium pH
- In the present study the effect of culture conditions were observed at different incubation time (1, 2, 3, 4, 5, 6 and 7 days), pH (5.0, 6.0, 7.0, and 8.5) and temperature (10˚C, 27˚C, 37˚C and 45˚C) and their biomass characteristics, biomass yield and protease productions were recorded.
2.5.2. Effect of Carbon and Nitrogen Sources
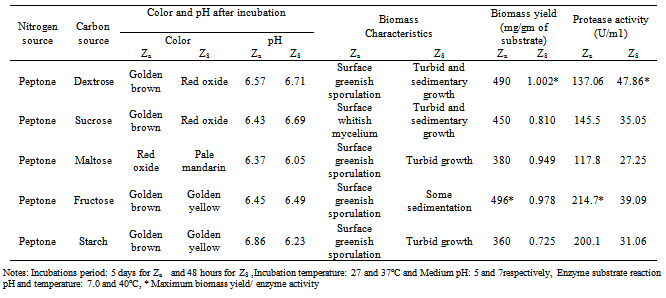
- For the production of extracellular proteases different carbon and nitrogen sources were studied in the liquid medium[15]. Sucrose, dextrose, fructose, starch or maltose were used as carbon source and peptone was used as nitrogen source to the medium and their effect on the production of protease, extracellular protein and biomass yield were recorded.
2.6. Optimum Conditions for Crude Enzyme Activity
- To determine the optimum conditions for the crude enzyme activity several factors such as pH (5.0 to 8.5), temperature (35 to 45˚C), incubation time (50, 60, 70, 80 and 90 minutes), Substrate concentration (i.e. 0.5, 1.0, 1.5, 2.0, 2.5 and 3.0%) and substrate specificity were observed.
3. Result and Discussion
- By using enrichment techniques five fungal isolates (ID as Za, Zb, Zc, Zd and Ze) and eleven bacterial isolates (ID as Z1 to Z11) were isolated from spoiled pulse and dry rotten shrimp respectively.
3.1. Screening and Identification of Selected Isolates
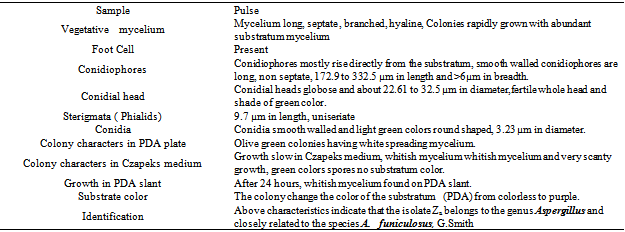
- All of these isolates were primarily screened by protein hydrolysis method and found that two fungal strains (Za and Zc) and two bacterial strains (Z8 and Z10) were vigorously hydrolyzed the egg albumin, skimmed milk casein and gelatin. Among them the fungal isolate Za and the bacterial isolate Z8 were the potent protease producers in the liquid medium that were finally selected for protease production. The selected fungal isolate was tested on the basis of their colony color, spore size, shape, arrangement, different types of conidiophores and sporangiophores, presence or absence of other special morphological features etc and compared with the standard description of “A manual of soil fungi”(Table.1)[20]. The fungal isolate was provisionally identified as Aspergillus funiculosus G. Smith (Fig.1.a, b)
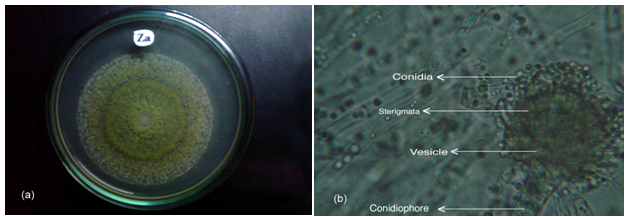 | Figure 1. The fungal isolate Za (A. funiculosus) (a) Colony of on PDA medium (b) Microscopic feature showing vegetative cells (10X12.5) |
|
|
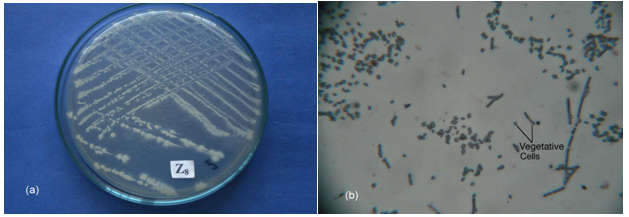 | Figure 2. The bacterial isolate Z8 (B. fastidiosus) (a) Colony on NA medium (b) Microscopic feature showing vegetative cells (40 X 12.5) |
3.2. Optimization of Different Cultural Conditions
- The present investigation was aimed to optimization of medium components which have been predicted to play a significant role in enhancing the production of proteases[22]. So, proper combination of various cultural conditions can be established for A. funiculosus and B. fastidiosus in order to suit for high secretion of protease.
3.2.1. Effects of Incubation Time
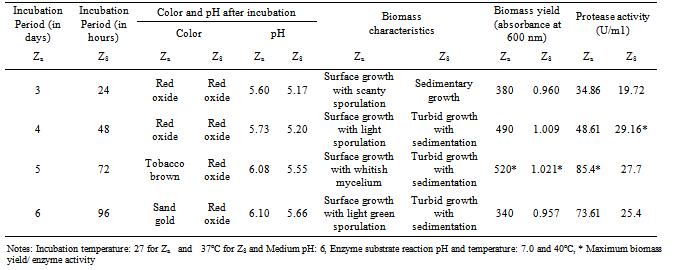
- In the present study the isolate A. funiculosus showed maximum activity (85.4 U/ml) after 5 days of incubation period which was similar with other research report[23]. On the other hand the bacterial isolate showed maximum protease activity (29.16 U/ml) after 2 days of incubation (Table.3).
|
3.2.2. Effects of pH
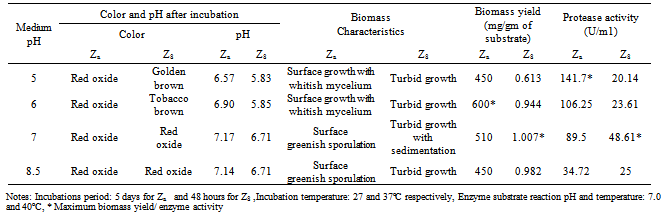
- Both of the isolates were allowed to grow in media of different pH ranging from 5.0 to 9.0. Maximum enzyme production was recorded at pH 5.0 and 7.0 by the isolates Za and Zc respectively (Table.4).
|
3.2.3. Effects of Temperature
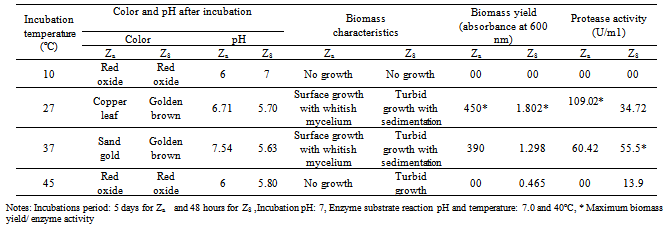
- Enzyme activity recorded at different temperatures revealed that the A. funiculosus yielded maximum protease production at 27ºC and B. fastidiosus at 37ºC (Table.5). Related studies also reported that protease production by Bacillus sp was best at 37°C[24,25]
|
3.2.4. Effects of Carbon and Nitrogen Sources
|
- Various carbon sources such as sucrose, fructose, maltose, starch and dextrose were used to obtain maximum protease production. When peptone was present as nitrogen source in growth media in that case fructose and dextrose increased the protease production compared to other carbon source for A. funiculosus and B.fastidiosus respectively (Table.6). Many authors reported variability of carbon and nitrogen sources with different organisms[26-32].
3.3. Factors Involved in Enzyme Activity
- Different factors such as pH, temperature, incubation time, substrate concentration play an important role for highest protease activity. So determination of these factors is necessary which help us to find out different limiting factor for maximum activity of proteases from microbial sources.
3.3.1. Effect of Reaction Time
- At 40ºC the crude enzyme of fungal and bacterial isolates showed highest protease activity when the reaction mixture were allowed to react for 70 and 60 minutes respectively (Fig. 4 a).
3.3.2. Substrate Specificity
- To observe the efficiency of protease activity during enzyme-substrate reaction different substrate such as casein, gelatin, bovine serum albumin (BSA) and egg albumin were used. Among this casein gave the highest value of activity with the crude of fungal and bacterial isolate (Fig.4 b).
3.3.3. Substrate Concentration
- Activity of protease has been shown to increase when the reaction mixture were allowed to react with different substrate concentration (0.5 to 3.0%). During enzyme-substrate reaction maximum protease activity was found in 1.5 % and 2.5 % casein for fungal and bacterial isolate respectively (Fig.4 c).
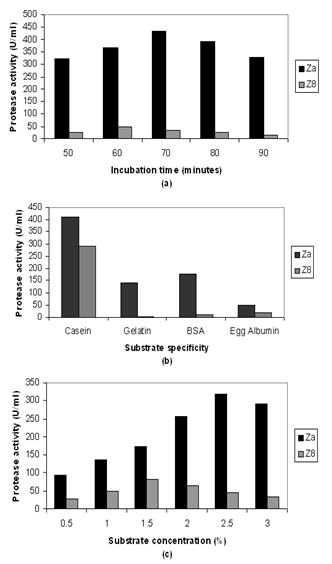 | Figure 4. Effects of various factors on the activity of protease of crude extract of fungal and bacterial isolate (a) Incubation time (b) Substrate specificity and (c) Substrate concentration |
3.3.4. Effect of Temperature and pH
- Protease activity increased with increase in temperature ranged from 35ºC to 45ºC. Maximum activity of protease (76.38 U/ml) was obtained at 45ºC with pH 7.0 by B. fastidiosus (Fig.5a). On the other handA. funiculosus showed highest enzyme activity (298 U/ml) at 35ºC with the reaction pH 6.0 (Fig.5b). Similar results at acid to neutral pH and different temperature was also reported by many authors[23,25, 27,33,34].
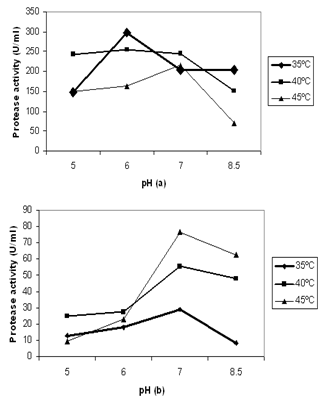 | Figure 5. Effects of temperature and pH on the activity of protease of crude extract of (a) fungal isolate and (b) bacterial isolate |
4. Conclusions
- The aim of this research work was to isolate and identify high protease producer from different sources. This might be an indication that the fungal strain A. funiculosus would produce sufficient amount of protease which could find applications in industry and biotechnology. Further work with the split of many other factor and interactions of each factors may provide clear picture about maximum production of protease by the selected isolates. In the present study all of the figures showed that the fungal isolate produced an increase amount of protease enzyme than the bacterial isolate and in case of both isolates it also revealed that protease activity was increased by specific factors. Further investigation on these two species can possibly reveal their potentiality as a source of protease for any biotechnological approach.
ACKNOWLEDGEMENTS
- Authors acknowledge the immense help received from the scholars whose articles are cited and included in references of their manuscript. The authors are also grateful to authors/editors/ publishers of all those articles, journals and books from where the literature for this article has been reviewed and discussed.
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML