-
Paper Information
- Previous Paper
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
Journal of Microbiology Research
p-ISSN: 2166-5885 e-ISSN: 2166-5931
2013; 3(1): 57-65
doi:10.5923/j.microbiology.20130301.09
Prevalence and Antibiotic Resistance Profiles of H. Influenzae and, S. pneumoniae Isolates from Clinical Samples of Patients in Mthatha, Eastern Cape Province, South Africa
Morobe IC 1, 2, Obi CL 1, 3, 2, Oyedeji AO 4, Vasaikar SD 1, Mbenza LB 2
1Department of Medical Microbiology, Faculty of Health Sciences, Walter Sisulu University, Nelson Mandela Drive, Mthatha 5117, Eastern Cape, South Africa
2Faculty of Health Sciences, Walter Sisulu University Nelson Mandela Drive, Mthatha 5117, Eastern Cape, South Africa
3Division of Academic Affairs and Research, Walter Sisulu University, Eastern Cape, South Africa
4Department of Chemistry, Faculty of Natural Sciences, Walter Sisulu University, Nelson Mandela Drive, Mthatha 5117, Eastern Cape, South Africa
Correspondence to: Morobe IC , Department of Medical Microbiology, Faculty of Health Sciences, Walter Sisulu University, Nelson Mandela Drive, Mthatha 5117, Eastern Cape, South Africa.
| Email: |  |
Copyright © 2012 Scientific & Academic Publishing. All Rights Reserved.
Haemophilus influenzae and Streptococcus pneumoniae are important causes of community acquired respiratory tract infections including, pneumonia, acute sinusitis, otitis media and meningitis, Pneumococcal infections are more common in the very young and very old individuals. The ability to effectively treat bacterial infections has been compromised in recent years due to the acquisition of antibiotic resistance, particularly to β-lactam drugs. The objective of the present study was to investigate the rate of isolation and antibiograms of H. influenzae and S. pneumoniae from clinical samples of patients in Mthatha, Eastern Cape Province, South Africa and the screening of antibacterial activity of medicinal plant extracts. Clinical samples were collected randomly from 289 individuals from different hospitals of Mthatha district, between February, 2010 and May 2011. H. influenzae and S. pneumoniae were isolated and positively identified by using morphological and biochemical tests. Antibiotic susceptibility tests were conducted on these pathogens using the agar diffusion test. MIC breakpoints were determined using E-test strips. A total of 289 patients were included in this study. From a total of 475 clinical samples tested, 323 (68.0%) were positive for both H. influenzae and S. pneumoniae. Most of the positive isolates were obtained from children under 9 years. Out of 323 isolates, 187(57.89%) were positive for H. influenzae and 136 (42.1%) were positive for S. pneumoniae. From 10 hospitals selected for sampling in this study, Mthatha General Hospital recorded the highest number of isolates, 42 (25.15%) and 31 (22.79%) of H. influenzae and S. pneumoniae positive isolates, followed by Nelson Mandela Academic Hospital 33 (19.76%). ST. Patricks 8 (4.79%) recorded the least number of isolates for H. influenzae, while Khotsong 4 (2.94%) recorded the least number of isolates for S. pneumoniae. Antibiotic susceptibility tests revealed Amoxicillin (MIC50,0.125μg/ml) and Vancomycin (MIC50,0.12μg/ml) as the most effective antibiotics against S. pneumoniae isolates and Co-amoxiclav (MIC50,0.3µg/ml) and Cefuroxime (MIC50,0.15µg/ml) against H. influenzae isolates. Cassine transvalensis showed the highest antibacterial MIC activity (0.16µg/ml and 0.32µg/ml), while Lycium inerme showed the least MIC activity (0.04µg/ml) for both organisms. These data highlight the need for education, training on vaccination programmes and to consider predominant resistance when choosing empiric therapies to treat bacterial infections.
Keywords: Antibiotics, Antibiotic Resistance, Infections, Susceptibility Test, Pathogens, Methanolic Extracts
Cite this paper: Morobe IC , Obi CL , Oyedeji AO , Vasaikar SD , Mbenza LB , Prevalence and Antibiotic Resistance Profiles of H. Influenzae and, S. pneumoniae Isolates from Clinical Samples of Patients in Mthatha, Eastern Cape Province, South Africa, Journal of Microbiology Research, Vol. 3 No. 1, 2013, pp. 57-65. doi: 10.5923/j.microbiology.20130301.09.
Article Outline
1. Introduction
- Pneumococcal infections are more common in the very young and very old individuals.S. pneumoniae or pneumococcus, is a Gram positive, alpha-hemolytic, bilesoluble aerotolerant aerobe and a member of the genus Streptococcus [1]. S. pneumoniae was recognised as a major cause of pneumonia in the late 19th century and was the subject of many humoral immunity studies [2]. The organism is an important cause of community-acquired respiratory tract infections including acute sinusitis, otitis media, meningitis [3]. The organism was first isolated in 1881 by the US Army physician George Sternberg and the French chemist Louis Pasteur, then known as the pneumococcus for its role as the etiologic agent of pneumonia [4]. The organism was termed Diplococcus pneumoniae from 1926 because of its characteristic appearance in Gram stained sputum [5]. It was renamed Streptococcus pneumoniae in 1974 because of its growth in chains in liquid media [5]. The ability to effectively treat pneumococcal infection has been compromised in recent years due to the acquisition of antibiotic resistance, particularly to β-lactam drugs [2].H. influenzae, formerly called Pfeiffer’s bacillus or Bacillus influenzae, is a small non-motile, non spore forming, Gram-negative rod shaped bacterium found in the upper respiratory tract of the host. It was first described in 1892 by Richard Pfeiffer during an influenza pandemic [5]. A member of the Pasteurellaceae family, it is generally aerobic, but can grow as a facultative anaerobe. H. infuenzae was mistakenly considered to be the cause of influenza until 1933, when the viral aetiology of the flu became apparent. Still, H. influenzae is responsible for a wide range of clinical diseases. H. influenzae was the first free-living organism to have its entire genome sequenced [6]. In 1930, two major categories of Haemophilus influenzae were discovered; the non encapsulated strains and the encapsulated strains. Encapsulated strains were classified on the basis of their distinct capsular antigens. The six generally recognized serotypes of encapsulated H. influenzae were a, b, c, d, e, and f. Genetic diversity among non encapsulated strains are greater than the capsulated group. Non encapsulated strains are termed non typeable (NTHi), because they lack capsular serotypes [7]. Non typeable H. influenzae (NTHi) are Gram-negative coccobacilli that asymptomatically colonize the nasopharynx, but cause respiratory infections such as bronchitis, otitis media, sinusitis, meningitis, epiglottitis, pneumonia, cellulitis, bacteremia and septic arthritis [7]. Standard tests to identify H. influenzae in clinical specimen X and V factor dependence have been reported [8], but do not distinguish between H. influenzae and a related non pathogenic species, H. haemolyticus. Such distinction has relied on the ability of H. haemolyticus to lyse horse red blood cells, which H. influenzae does not do. However, this phenotype may be lost during passage[8]. The increasing global prevalence of antibiotic resistance among H. influenzae and S. pneumoniae caused a great concern and necessitated the development of rapid and sensitive DNA-based assay to improve the accuracy of the diagnosis of S. pneumoniae and H. influenzae infections [4, 9, 10, 11, 12, 13, 14].. Until recently, β-lactamase production has been the primary mechanism of antibiotic resistance in H. influenzae and S. pneumoniae. As a result of the rapid evolution of β-lactam resistant S. pneumoniae, macrolides have been increasingly used as initial empirical therapy in community acquired infections. However, the global increase in microlide-resistant strains of S. pneumoniae now threatens to compromise the use of these antibacterials for the treatment of these conditions[15].Severe pneumonia and meningitis have been reported to respond to chloramphenicol and benzylpenicillin and mild pneumonia to trimethoprim-sulphamethaxazole, ampicillin or amoxicillin [1]. However, misuse of antibiotics for respiratory tract infections in children is widespread and fuelled by public attitude and expectations heralding the emergence of resistance. Resistance levels of up to 84% for co-trimoxazole, 52% for penicillin, and 25% for ampicillin were reported [1].Resistance of bacteria to antibiotics is reported to depend on serotypes, geographical, seasonal and clinical factors, making susceptibility profiles to vary with time and geographical areas. Such resistance trends highlight an urgent need for new antibacterials for treatment of infections [16, 17, 18 ]. Currently, there is a continuous spread ofmulti-resistantpathogens which have become a serious threat to public health and infection control practices worldwide [1]. This problem has necessitated a search for new antimicrobial compounds from other sources including plants. The antimicrobial compounds derived from plants that can either inhibit the growth of pathogens or kill them and have no or least toxicity to host cells are considered candidates for developing new antimicrobial drugs. It is expected that plant extracts showing target sites other than those used by antibiotics will be active against drug resistant microbial pathogens [19].This investigation was carried out to unravel the prevalence antimicrobial resistance profiles and screening of Antibacterial Activity Plant Extracts of H. influenzae and S. pneumoniae isolates from clinical samples of patients in the Nelson Mandela Academic Hospital Complex, Mthatha General Hospital and other Satellite hospitals in Mthatha, Eastern Cape, South Africa.
2. Materials and Methods
2.1. Sampling
- A prospective study based on laboratory investigations at the microbiology laboratory of the National Health Laboratory Services (NHLS-Mthatha) at Nelson Mandela Academic Hospital and Department of Medical Microbiology, Faculty of Health Sciences, Walter Sisulu University was undertaken.In this study 475 samples were randomly obtained from patients between February, 2010 and May 2011. The samples were collected from the Nelson Mandela Academic Hospital (NMAH) and Mthatha General Hospital. Majority of the samples were from the Mthatha General Hospital (MGH).The remaining samples were received from surrounding Satellite hospitals such as Bedford Orthopedic, Madwaleni, Saint Elizaberth, All Saints, Khotsong, Canzibe, Mount Ayliff and Saint Patricks. Samples collected included; blood cultures, pus, ear swab, throat swabs, cerebrospinal fluid (CSF) and sputum cultures. H. influenzae, S. pneumoniae, S. aureus and S. pyogenes were isolated from the specimens and identified down to the species level using standard microbiological procedures (CLSI 2010). Antimicrobial susceptibility testing was conducted on these isolates (pathogens) using the modified Kirby-Bauer disc agar diffusion test and MIC breakpoints were determined using the E-test strips.
2.2. Enrichment, Culturing, Morphological and Biochemical Identification
- On arrival at the laboratory, samples were directly streaked onto blood and chocolate agar (Merck, Darmstadt, Germany) and then incubated at 37°C in an enriched CO2 incubator for 24 h. α-hemolytic colonies appear when S. pneumoniae is grown on blood agar overnight under aerobic conditions at 37°C. A Gram stain of suspected colonies from the media was made and Lancet shaped Gram positive diplococci or cluster-forming colonies was observed. Growth of these bacteria is inhibited by low concentrations of the surfactant Optochin and the cells are lysed by bile acids. Bacterial culture of H. influenzae was performed on blood and Chocolate agar, with X (Hemin) and V (NAD) factors added on Mueller Hinton agar (Merck, Darmstadt, Germany) and incubated at 37°C for 24 h in an enriched CO2 incubator. Colonies of H. influenzae appear as convex, smooth, pale, grey or transparent colonies. Gram- stained and microscopic observation of a specimen of the organism showed Gram negative, coccobacilli, with no specific arrangement. Suspected colonies were subcultured onto blood and chocolate agar (Merck, Darmstadt, Germany) slants were maintained at 4℃. Following these, the catalase test was performed on all isolates as previously described (Morobe et al., 2009). Isolates that were confirmed as S. pneumoniae and H. influenzae were preserved in a solution containing 80% tryptose soy broth (Oxoid) and 20% glycerol at -80°C for use in the steps that followed.
2.3. Antibiotic Susceptibility Testing
- Isolates that were confirmed as H. influenzae and S. pneumoniae were inoculated on Mueller Hinton broth (Merck, Darmstadt, Germany). The flasks were incubated at 37℃ on a Gallenkamp shaker (200 rpm) for 24 h. The turbidity of the actively growing broth culture was adjusted with sterile saline to obtain turbidity optically comparable to that of the 0.5 McFarland standard. One millilitre of the cell suspension was then transferred onto the surface of blood and chocolate agar (Merck, Darmstadt, Germany) and then spread evenly. The susceptibilities of all isolates to different antimicrobial agents were tested by disc-agar gel method as standardized by the National Committee for Clinical Laboratory Standards (NCCLS), [2010].The following panel of antimicrobial discs and concentrations were used; Ampicillin (30μg), Penicillin G (30μg), Amoxicillin (30μg), Fucidic acid (30μg), Rifampicin (30μg), Ciprofloxacin (5μg), Tetracycline (30μg), Cotrimoxazole (30μg), Cefotaxime (30μg), Coamoxiclav (10μg), Cefuroxime (30μg),Erythromycin (5μg), Vancomycin (30μg) and Clindamycin (2μg), Cloxacillin (30μg), Imipenem (30μg) (Mast Diagnostics, Merseyside, UK). The reference strains of both H. influenzae and S. pneumoniae were obtained from the South African Bureau of Standards (Pretoria, South Africa). The MIC breakpoints for antibiotics were obtained optically using the E-test strips [4]. Unlike antibiotic discs, the E-test strip was placed onto the blood agar plates and the MIC was determined by observing the point where the zone of inhibition ended on the agar plate.
2.4. Screening of Antibacterial Activity of Plant Extracts
2.4.1. Disc Diffusion Assay
- The disc diffusion technique as previously described [20], was used to test for antimicrobial activity. Mueller-Hinton broth culture of each isolate was standardized to 0.5 McFarland standard (i.e. 1x106 CFU/ml). The bacterial suspension was spread over the Mueller-Hinton agar. The paper disc impregnated with 15μl Croton grattissimus, Lycium inerme, Cassine transvalensis and Vanguera infuesta plant extracts was placed onto the plate that was spread with the bacterial suspension. The plate was incubated at 37°C overnight. The positive results were indicated by the clear zone around the disk [20].
2.4.2. Serial Dilution Assay for Determination of the Minimal Inhibitory Concentration (MIC)
- A micro-dilution technique using a sterile 96 well micro plates, as described by [21], was used to obtain MIC values of the crude extracts against the bacteria that tested positive in the above method. The microbial cultures were diluted in fresh Nutrient broth to a 0.5 McFarland standard (approximate inoculum size of 1x106 CFU/ml) and 100 µl from each sample was added to all wells. Each plant extract was serially diluted to obtain 2.5 mg/ml starting concentration in the first well. Similar serial dilutions were performed for Ampicillin (1mg/l), as a positive control obtained from Sigma. The starting concentration in the first well after the dilution was 0.25 mg/ml. An equal volume of 100 µl fresh bacterial culture was added to the wells. Micro plates were covered with lids and incubated at 37℃ overnight. P-Iodonitrotetrazolium (INT) (Sigma) reagent (0.2 mg/ml) was used (40 µl/well) to indicate the presence of uninhibited bacterial growth (a pink/purple colour) or the inhibition (colourless) of bacterial growth in each well. The lowest concentration of the extract that was inhibitory was taken as the MIC of a crude extract. Only extracts that showed the antibacterial activity from the disc diffusion assay were tested for MIC [22].
2.4.3. Minimum Bactericidal Concentration (MBC) of the Extracts Against the Test Organisms
- From the tubes showing no visible sign of growth in MIC determined, 0.1ml was inoculated onto sterile chocolate agar plates by the spread plate method. The plates were then incubated at 37℃ for 24 h. The least concentration that did not show growth of test organisms was considered as the MBC.
3. Results
3.1. Prevalence of H. influenzae and S. pneumoniae
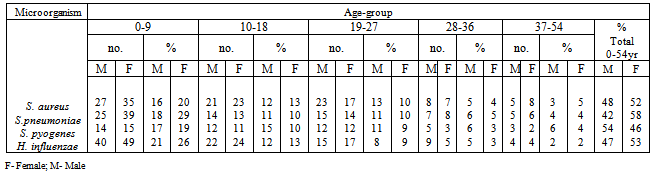
- From a total of 475 clinical samples tested, 323 (63.79%) were positive for both H. influenzae and S. pneumoniae. Out of 323 isolates, 187 (57.89%) were positive for H. influenzae and 136 (42.1%) were positive for S. pneumoniae. From 10 hospitals selected for sampling in this study, Mthatha General Hospital recorded the highest number of isolates, 42 (25.15%), 31 (22.79%) respectively for H. influenzae and S. Pneumonia, followed by Nelson Mandela Academic Hospital 33 (19.76%) and 26 (19.12%) respectively. While ST. Patricks 8 (4.79%) recorded the least number of isolates for H. influenzae and Khotsong 4 (2.94%) recorded the least number of isolates for S. pneumoniae as shown in table 1 and figure1.Table 2, depicted the prevalence of the isolates in different age-groups. From this results the age group 0-9 years recorded the highest number of positive isolates (19.5%) from males and 27.5% from females), while the age group 37-54 years recorded the least number of isolates (3.0%) from both males and females (p < 0.05). From this results females recorded the highest total number of positives isolates from different age groups (55.5%) than males (44.5%).
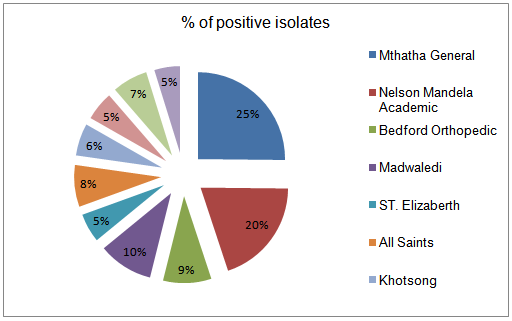 | Figure 1. Percentage of positive isolates from different hospitals |
|
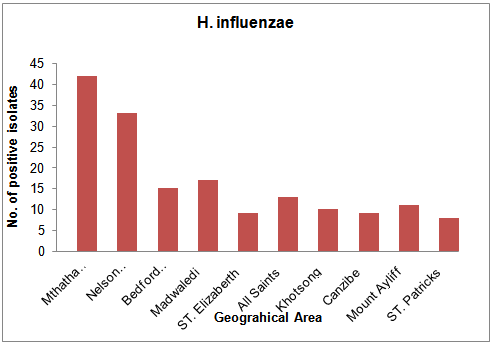 | Figure 2. Geographical distribution of H. influenzae |
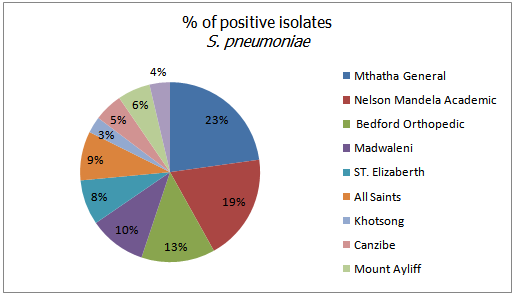 | Figure 3. Percentage of positive isolates of S. pneumonia |
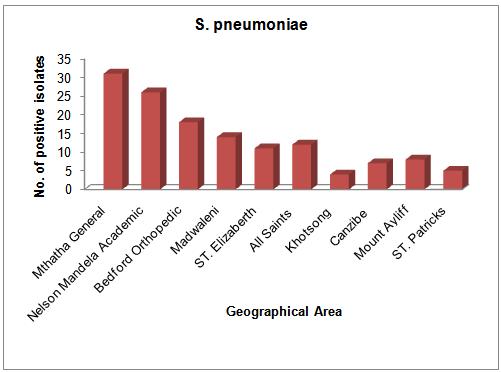 | Figure 4. Geographical distribution of S. pneumoniae |
3.2. Antimicrobial Susceptibility Testing of H. influenzae and S. pneumoniae
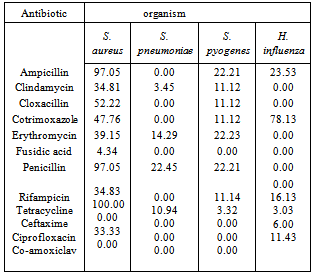
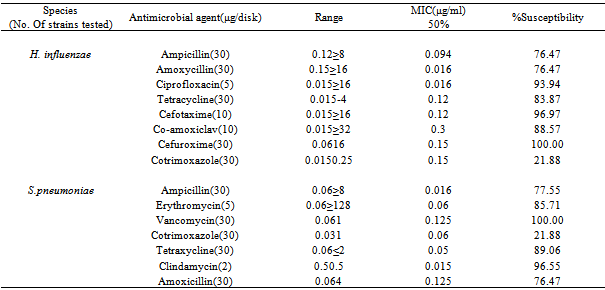
- Penicillin G resistance rates for H. influenzae and S. pneumoniae were 0.00% and 22.45% respectively (Table 3). Antibiotic susceptibility test revealed Ceftaxime (97.07%) and Co-amoxiclav (88.57%) were the most effective antibiotics against the isolates.
|
3.3. Effects of Methanolic Extracts on Bactericidal Activity
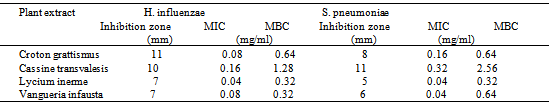
- In this study, crude extracts were derived from the plant bark and leaves and their efficacy to inhibit the growth of multidrug resistant pathogens (H. influenzae and S. pneumoniae) were studied. The results presented in table 5 indicated that methanolic extracts inhibited most strains of S. pneumoniae (24.26%) than H. influenzae (11.23%).The plant extracts showed antibacterial activity against the isolates with zone of inhibition ranging from 2 to 16mm (tables 5). The MIC of the plant extracts ranged from 0.04μg/ml to 0.16μg/ml for H. influenzae, while that of S. pneumoniae ranged from 0.04μg/ml to 0.32μg/ml for (table 5). Cassine transvalensis showed the highest antibacterial MIC activity (0.16µg/ml and 0.32µg/ml), while Lycium inerme showed the least MIC activity (0.04µg/ml and 0.04µg/ml) for both organisms.
|
|
4. Discussion
- H. influenzae and S. pneumoniae are known to cause community acquired respiratory tract infections including, pneumonia, acute sinusitis, otitis media, meningitis, and bacteremia, in various age groups. Pneumococcal infections are more common in the very young and very old individuals. The ability to effectively treat bacterial infections has been compromised in recent years due to the acquisition of antibiotic resistance, particularly to β-lactam drugs. The present study investigated the occurrence and antibiograms of H. influenzae and S. pneumoniae from clinical samples of patients in Mthatha district, OR Tambo Manucipality, Eastern Cape Province, South Africa. The results in this study revealed that, there were no significant difference in the prevalence of the pathogens in the various hospitals and with respect to sexes (p > 0.05), suggesting that respiratory tract infections is not hospital or sex linked. Poor hygienic conditions and other factors may enhance the spread of pathogens. The number of isolates recorded in various age groups showed that there was significant difference (p < 0.05) in the infection rate, for instance, 0-9 years recorded the most number of positive isolates (19.5% from males and 27.5% from females), while the age group 37-54 years recorded the least number of isolates (3.0% from both males and females). In another study (Weiss et al., 2001), results obtained were different from the current study, where S. pneumoniae was the most frequently incriminated bacterial pathogen among the designated pathogens.These results correlate with the data previously reported [16], where 53% of females between the age of 0-5 years were infected with H. influenzae and S. pneumoniae, where a significant difference was indicated in total percentage recorded for each age group to [24,1] results, where the highest infected age groups of 0 to 9 was 84% and the least infected age group of 73 to 81 (3.0%).The difference in the above results may have been due to the size of the samples, time, geographical distribution and seasonal variations. Antibiotic susceptibility test revealed Amoxicillin (MIC50, 0.125μg/ml) and Vancomycin (MIC50, 0.12μg/ml) as the most effective antibiotic against S. pneumoniae isolates, while Co-amoxiclav (MIC50, 0.3µg/ml) and Cefuroxime (MIC50, 0.15µg/ml) were effective against H. influenzae isolates. In previous studies in USA [4, 25], the antibiotic susceptibilities for both organisms were similar to the present study (MIC50, 0.12 or 0.125µg/ml) and lower than previously reported findings [23], MIC50 ≥4µg/ml).The high rates of resistance to Penicillin G (47.05%) and Ampicillin (47.33%), in contrast with the marked levels of susceptibilities of isolates to Clindamycin (16.33%) and Cloxacillin(21.11%), which are less frequently used. In a similar study conducted in France [9], similar results were obtained.Thus suggesting the relationship between antibiotic use and the level of drug resistance encountered in this study. In fact, most of our isolates demonstrated high levels of resistance to Ampicillin and Penicillin G. This results correlate with the previous findings [1]. Plant extracts used in this study exhibited antibacterial activity against both pathogens tested. S. pneumoniae was the most susceptible organism (24.26%) to the methanolic extracts. Gram negative bacilli (H. influenzae) demonstrated varying levels of susceptibility in terms of zone of inhibition diameters, which ranged from 2-16mm. However, only four plant extracts (Cassine transvalensis, Croton grattisimus, Lycium inerme, and Vangueria infausta) showed a high potency MIC (0.04 – 2.56 mg/ml) on all tested isolates. However, There was a significant difference between zones of inhibition diameters of all four extracts for each organism. This may be due to interactions between factors related to pathogens, the environment, the rate of diffusion of the extract, culture medium and depth of the medium. The high activity levels in Gram positive cocci when compared to Gram negative bacilli, could be explained by the different cell wall structures of these bacteria. Gram negative bacteria possess an outer phospholipid membrane with structural lipopolysaccharide components that are not found in Gram positive bacteria. This composition makes the cell wall impermeable to lipophilic solutes and the porins in the cell wall do not allow the penetration of high molecular mass hydrophilic solutes [25].The activity of the methanolic extracts were compared with that of commercial antibiotic disc Comparing the diameters of zone of inhibition obtained from the commercial antibiotics, both organisms were resistant to Penicillin G and Ampicillin of the eleven antibiotics tested. They were susceptible to clindamicin and cloxacillin. Despite methanolic plant extracts being active against these respiratory tract bacterial pathogens, their activity is lower than that of the commercial antibiotics. However, the wide spectrum of activity of plant extracts compared to the commercial antibiotics is an indication of their antibacterial potential in medicine. There was no significant difference in the MBC of the extracts for both H. influenzae and S. pneumoniae. The differences observed could be attributed to the fact that serotypes of pneumococci vary geographically and are subject to antigenic changes. This study confirms the traditional antimicrobial use of the four (Cassine transvalensis, Vangueria infuesta, Croton grattisimus and Lycium inerme) tested plant species in bacterial respiratory tract infections.
5. Conclusions
- These data highlight the need for Education, training on vaccination programmes and to consider predominant resistance when choosing empiric therapies to treat bacterial infections. It also demonstrated that medicinal plant extracts could be sources of compounds which might be useful in managing respiratory tract infection pathogens. However, further studies about the isolation of active compounds, are important to better options to some of the antibiotics commonly used for respiratory tract infections in the environment and propose the use of plant extracts as alternative approaches to resistance management.
ACKNOWLEDGEMENTS
- The authors wish to thank Walter Sisulu University, NRF and MRC for financial assistance. We are indebted to the technical staff of the Department of Medical Microbiology, Walter Sisulu University (WSU) for the technical assistance they provided during this research work. Special thanks go to the management and technical staff of the National Health Laboratory Services, Nelson Mandela Academic Hospital, Mthatha and The Laboratory for Emerging and Infectious Diseases, Tohoku University, Japan for the outstanding technical assistance provided during this research work.
References
| [1] | Ndip RN, Ntiege EA, Ndip LM, Nkwelang G, Akoachere TK, Akenji A (2008). Antimicrobial resistance of bacterial agents of the upper respiratory tract of school children in Buea, Cameroon. J. Health, Pop. and Nut. 26:397-404. |
| [2] | Rosen JB, Thomas AR, Lexan CA, Reingold A, Hadler JL, Harrison LH., 2011.Geographic variation in Invassive Pneumococcal Disease Following pneumococcal conjugate vaccine introduction in the United States. Clin. Infect. Dis. 53: 453-456. |
| [3] | Sunakawa K, Farrell DJ (2007). Mechanism, molecular and sero-epidemiology of antimicrobial resistance in bacterial respiratory pathogens isolated from Japanese children. J. Clin. Microbiol and Antimicrob. 6:1476-1489. |
| [4] | Doern GV, Jones RN, Pfaller MA, Kukler K (1998). Haemophilus influenzae and Morexella catarrhalis from patients with community-acquired respiratory tract infections. J. Antimicrob. Agents. Chemother. 43:385-389. |
| [5] | Turk DC (1984). The pathogenicity of Haemophilus. J. Med. Microbiol. 18: 1- 6. |
| [6] | Meats E, Feil EJ, Stringer S, Cody AJ, Goldstein R, Kroll JS, Popovic T, Spratt BG (2003). Characterization of Encapsulated and Noncapsulated Haemophilus influenzae and Determination of Phylogenetic Relationships by Multilocus Sequence Typing. J. Clin. Microbiol. 41:1623-1636. |
| [7] | Chang CM, Lauderdale TL, Lee HC, Lee NY, Wu CJ, Chen PC, Ko WC (2010). Colonization of fluoroquinolone resistant Haemophilus influenzae among nursing home residents in Southern Taiwan. J. Hospital Infect. 75: 304-308. |
| [8] | Kilian (2005). Pathogenetic species of the genus Haemophilus and Streptococcus pneumoniae produce immunoglobulin A1 protease. J. Infect. Imm. 26: 143-149. |
| [9] | Hoban DJ, Doern GV, Fluit AC, Roussel-Delvallez M, Jones RN (2001).Worldwide prevalence of antimicrobial resistance in Streptococcus pneumoniae, Haemophilus influenzae and Moraxella catarrhalis in the SENTRY antimicrobial surveillance program, 1997-1999. J. Clin. Infect. Dis. 32:1058-4838. |
| [10] | Akoachere, TK, Ndip RN, Chenwi EB, Ndip LM, Njock TE, Anong (2002). Antibacterial effect of Zingiber officinale and Garcinia kola on respiratory tract pathogens. East African Med. J. 79:11. |
| [11] | Bennett D, Lennon B, Humphreys H, Cafferkey M (2003). Penicillin susceptibility and epidemiological typing of invasive pneumococcal isolates in the Republic of Ireland. J. Clin. Microbiol. 47:3641-3648. |
| [12] | Turnidge JD, Bell JM (2009). The SENTRY Asia-Pacific group: Emerging beta-lactamase negative ampicillin resistant Haemophilus influenzae in Japan and South Africa. J. Antimicrob. Agents and Chemother. 43: 136. |
| [13] | Dias R, Canica M (2004). Emergence of invasive erythromycin-resistant Streptococcus pneumoniae strains in Portugal; Contribution and phylogenetic relatedness of serotype 14. J. Antimicrob. Chemother. 54:1035-1039. |
| [14] | Ghalem BR, Mohamed B (2009). Essential oil from gum of Pistacia atlantica Desf for screening of antimicrobial activity. African J. Pharm. and Pharmacol. 3:087-091. |
| [15] | Fani F, Leprohon P, Legari D and Quellette M., 2011. Whole genome sequencing of penicillin- resistant Streptococcus pneumonia reveals mutations in penicillin binding proteins and in a putative iron permease. Genome Biol. 12:115. |
| [16] | Jones RN, Farrell DJ, Morrissey I (2003). Quinupristin - Dalfopristin resistance in Streptococcus pneumoniae ; Novel L22 Ribosomal protein mutation in two clinical isolates from the SENTRY antimicrobial surveillance program. J. Antimicrob. Agents Chemother, 47: 2696-2698. |
| [17] | Morrissey I, Farrell DJ, Bakker S, Buckridge S, Felmingha (2003). Molecular characterization and antimicrobial susceptibility of fluoroquinolone-resistant or susceptible Streptococcus pneumoniae from Hongkong. J. Antimicrob. Agents Chemother. 47:1433-1435. |
| [18] | Iwalokun BA, Ogunledun A, Ogbolu DO, Bamiro SB, Juni-Omojola J (2004). In vitro antimicrobial properties of aqueous garlic extract against multidrug-resistant bacteria and Candida species from Nigeria. J. Med. Food. 7: 327-333. |
| [19] | Rajendran NK, Ramakrishnan J (2009). In vitro evaluation of antimicrobial activity of crude extracts of medicinal plants against multi-drug resistant pathogens. J. Med. Microbiol. 2: 97-101. |
| [20] | Samie, A, Housein, A, Lall, N, and Meyer, JJM, 2009. Crude extracts of purified compounds from Pterocarpus angolensis and the essential oil of Lippia against selected bacteria and Entamoeba histolytica. Annals of Tropical Medicine and Parasitology, 103: 427-439. |
| [21] | Eloff JN, Lwalewa EO, Suleiman MN, Mdee LK (2009). Antifungal and antibacterial activities of different extracts of Harungana madagascariensis stem bark. Journal of Drug Assessment, 7: 64-78. |
| [22] | Mathebe, MC, Nikolova, RV, Lall, N, Nyazema, NZ, 2006. Antibacterial activity of medicinal plants used for the treatment of diarrhoea in Limpopo Province, South Africa. Journal of Enthnophramacology, 105: 286-293. |
| [23] | Weiss K, Restieri C, Gauthier R, Laverdiere M, McGeer A, Davidson RJ, Kilburn L, Bast DJ, de Azavedo J, Low DE (2001). A nosocomial outbreak of fluoroquinolone-resistant Streptococcus pneumoniae. J. Clin. Infect. Dis. 33:517-522. |
| [24] | Greenberg D, Dagan A, Muallem M, Porat N (2003). Antibiotic-resistant invasive pediatric Streptococcus pneumoniae clones in Israel. J. Clin. Microbiol. 41:5541-5545. |
| [25] | Thornsberry C, Sahm DF, Kelly LJ, Critchley IA, Jones ME, Evangelista AT, Karlowsky JA (2002). Regional trend in antimicrobial resistance among clinical isolates of Streptococcus pneumoniae, Haemophilus influenzae, and Moraxella catarrhalis in the United States. J. Clin. Infect. Dis. 34: 4-16. |
| [26] | Douglas M, Bennett’s (2000). Princ. and Pract. Infect. Dis. Fifth Edition, Volume 2, 2369-2376. |
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML