Odeyemi A. T. , Ajayi A. O , Igbalajobi O. A.
Microbiology Department, Ekiti State University, Ado-Ekiti, Ekiti State, 36001, Nigeria
Correspondence to: Odeyemi A. T. , Microbiology Department, Ekiti State University, Ado-Ekiti, Ekiti State, 36001, Nigeria.
| Email: |  |
Copyright © 2012 Scientific & Academic Publishing. All Rights Reserved.
Abstract
This research determined the microbial counts, antibiotics susceptibility and plasmid profile of the isolated bacteria of water samples from Arinta waterfall Ipole-Iloro Ekiti, Ekiti State, Nigeria. The total bacterial and coliform counts were done using pour plating technique while the susceptibility of the isolated microbes were determined using disc diffusion technique the plasmid profile of the isolates was determine using alkaline lysis method and agar gel electrophoresis. The mean total bacterial count of the water sample ranged 106 X 102 – 15.5 X 103 CFU/ml, while the mean coliform count ranged 97.5 X 102 – 6.50 X 103 CFU/ml. The identified bacteria isolates and their percentage distribution are Escherichia coli which have the highest percentage (31.1%), Klebsiella spp. (22.2%), Pseudomonas spp. (8.89%), Citrobacter spp. (6.67%), Salmonella spp. (6.67%), Serratia spp. (6.67%), Bacillus spp. (4.44%), Aeromonas spp. (4.44%), Micrococcus spp. (4.44%) and Aerococcus spp. (4.44%). Among these ten species, multiple antibiotics resistant bacteria were identified. The most active antibiotics were ofloxacin (14%), ciprofloxacin (28%) and gentamicin (17%). 99.6% of the isolates exhibited multiple antibiotic resistances. Eight bacterial isolates harbored plasmids all of which were not less than 2.51kbp in molecular weight. Hence, it showed that the gene coding for the isolates were located on the plasmid DNA while the remaining isolates which have no plasmid might showed gene coding for antibiotic resistance being located on chromosomal DNA. The occurrence of plasmid mediated multidrug resistant in bacteria in this surface water lightens the public health concern.
Keywords:
Plasmid Profile, Coliform, Multiple Antibiotic Resistance, Arinta Waterfall
Cite this paper: Odeyemi A. T. , Ajayi A. O , Igbalajobi O. A. , Plasmid Profile of isolated Bacteria from Arinta Waterfall in Ipole-Iloro Ekiti, Journal of Microbiology Research, Vol. 3 No. 1, 2013, pp. 32-38. doi: 10.5923/j.microbiology.20130301.05.
1. Introduction
Water is essential to life and it is of fundamental importance to all living organisms and constitutes about 90% by weight of the human body[1]. The availability of good drinking water is an important ingredient for preventing epidemic disease and improving the quality of life[2]. In developing countries, access to both clean water and sanitation are of less concern, and waterborne infections are common. Two and a half billion people have no access to improved sanitation, and more than 1.5 million children die each year from diarrheal diseases[3]. According to the world health organization[4], 80% of all diseases are attributed to unsafe water with about 1 ¼ billion people in the world suffering from major related disease at any time. In recent years, a tremendous amount of attention has been directed toward pollution of soil and water supplies and the subsequent effects on the life of many animals and human. Meanwhile, pollution of the environment simple entails making the environment unclean and unhealthy by aiding unwholesome states or condition[5].The occurrence and spread of antibiotic-resistant bacteria are pressing public health problems worldwide, and aquatic ecosystems are a recognized reservoir for antibiotic-resistant bacteria and antibiotic resistance genes[6]. Naturally occurring antibiotic-resistant bacteria and antibiotic resistance genes in the aquatic environment are selected for and enriched for by antibiotics found in sewage and agricultural runoff, which result from the widespread and increased use of antibiotics[7]. Historically, concerns about the microbial quality of drinking water have focused on the occurrence of pathogens in drinking water distribution systems[8].The research findings focused on determining the bacterial load of the water samples from Arinta waterfall in Ipole - Iloro Ekiti. The antibiotic susceptibility and the plasmid profile of the isolated microbes were determined to evaluate whether the gene coding for the resistance is located on the chromosome or plasmid.
2. Materials and Methods
2.1. Study Area
Arinta waterfall in Ipole-Iloro Ekiti was located at about 6km Northwest of Ikogosi Ekiti in Ekiti State, between latitude 70451 to 8051 North of the equator and longitude 40451 to 50451 East of the Greenwich Meridian. The fall cascades down the rocky hills from about 50m height (Figure 1). | Figure 1. Arinta water fall, Ipole-Iloro Ekiti under the influence of anthropogenic activities |
2.2. Collection of Samples
The water samples were collected from seven different points in which one is directly from the point where the waterfall and the other six from different point along the flowing path of the waterfall about 120mm away from each other with the use of sterile sampling bottles. Water samples were collected in triplicates according to the Standard Methods for the Examination of Water and Wastewater[9]. The collected samples were kept at 4℃ in the ice parked box and transported to the laboratory for immediately analysis within 4 h of collection.
2.3. Isolation and Characterization of Microbes
In all cases, water samples were shaken vigorously, and dilutions were prepared in sterile saline (0.9 g NaCl/100ml of sterile distilled water). Triplicate plates were used for each sample dilution. The original numbers of organisms (CFU / ml) in water samples were calculated considering the dilution factor and the final estimate was taken as the average of figures obtained from the countable plates[9]. After preparation of the serial dilution of the water samples above 10-4 one millimeter of each dilution 10-2 and 10-3 were drawn aseptically from each dilution tubes and inoculated into sterile Petri dishes. The media were then added to the plates after cooling to about 45℃. Each plates of the same sample were known to contained nutrient agar for one, MacConkey agar and Eosin methylene blue for the other. The bacterial isolates were identified as described by[10] and pure cultures of isolates were kept on nutrient agar slants at 12℃ until used.
2.4. Antibiotics Sensitivity
Antibiotic susceptibility of isolates was determined using disc diffusion method on Mueller – Hilton agar according to CLSI guidelines[11]. The antibiotic multi-disc containing augmentin(AUG, 30ug), ampicillin(AMP, 25ug),cotrioxazole (COT, 25ug), nalidixic acid(NAL, 30ug), nitrofurantoin(NIT, 300ug), tetracycline (TET, 25ug), erythromycin (ERY, 15ug), penicillin (PEN, 10u), chloramphenicol (CHL, 30ug) (Abtek Biologicals Ltd, UK) were used.Four pure colonies of each isolate on a 24 h plate culture were randomly selected and inoculated into 2 mL of sterile peptone water broth in bijou bottles. This was incubated at 37℃ for 6 h and the turbidity was adjusted by serial dilution in phosphate buffer saline (pH 7. 2) to match an opacity tube containing 0.5 ml of 1% Barium Chloride in 1% Sulphuric acid (McFarland’s 0.5 Barium Sulphate standard containing 105 CFU/ml of the inoculums).One milliliter of the culture dilution (bacteria suspension) was transferred into a well dried surface of diagnostic sensitivity test agar (DST) medium and tilted to spread evenly over the entire surface of the agar plate. The excess fluid was drained off and dried within 5 min multi-antibiotic discs were then placed of the surface of the inoculated plate and incubated aerobically at 37°C for 18 to 24 h (over-night). The diameter of the zone of inhibition was measured in millimeter and was reported as susceptible or resistant when compared with antibiotics chart.
2.5. Plasmid Profile
2.5.1. Detection of Plasmids Among Antibiotic-resistant Isolates
Plasmid analysis was performed on representative isolates selected on the basis of their antibiotic resistance phenotypes. All isolates selected for plasmid analysis were multiple resistant isolates with diverse phenotypes. The modified alkaline lysis method for plasmid extraction described by[12] was used for extraction of plasmid.
2.5.2. Extraction of Plasmid
Organisms were grown in 2.5ml of nutrient broth and incubated at 35℃ for 18h. After incubation, 0.5 ml of each culture was transferred into 1.5ml Eppendorf tubes for plasmid extraction and glycerol was added to the remaining culture and stored at 4℃. The Eppendorf tubes were centrifuged at 6,000rev/min for 15 seconds after which the supernatant was carefully removed with the use of fine-tip automatic micropipette and the cell pellet was thoroughly suspended in the 100µl of lysozyme solution. The pellet-lysozyme mixture was incubated at 0℃ for 30mins after which 200ml of the alkaline sodium doedecyl sulphate (SDS) solution was added and gently vortexed. This suspension was almost clear and slightly viscous. The tube was maintained for 5mins at 0℃ and then 150µl of sodium acetate solution was added. The content of each tube was gently mixed for about six to seven seconds during which clots of DNA were observed in each tube. The tubes were maintained at 0℃ for 60mins to allow most of the protein, high molecular weight RNA and chromosomal DNA to precipitate. The tubes were centrifuged for 5mins at 15,000rev/minute to yield a clear supernatant. About 0.4ml of the supernatant was removed from each tube and transferred into smaller centrifuge tubes. One millilitre of cold ethanol was added and held at -20℃ for 30Omins. The precipitate was then collected by centrifugation at 6000 rev/min for 2mins and the supernatant was removed by aspiration. The pellet was dissolved in 100µl of 0.1M sodium acetate/0.05 Tris HCl (pH 8) and reprecipitated in 2vols of cold ethanol. After 10 min, at -20℃, the precipitate was again collected by centrifugation as described earlier. The pellet was dissolved in 40µl of water and then 10µl of sample buffer was added. Between 10-20µl of plasmid DNA in solution was applied to an agarose gel for electrophoresis.
2.5.3. Agarose Gel Electrophoresis
One percent agarose was prepared and loaded into electrophoresis chamber containing between 12-18 wells. The electrophoresis buffer that was used contained 40mM Tris, 20mM sodium acetate, 2mM EDTA, adjusted to pH 7.8 with acetic acid. The sample buffer contained 25% sucrose, 5mM sodium acetate, 0.05% bromophenol blue and 0.1% SDS. Electrophoresis was allowed to proceed at room temperature until bands become visible at the positive end of the chamber. After electrophoresis, gels were stained with ethidium bromide (1µl/ml) and viewed under UV trans illumination. The molecular marker that was used was the bacteriophage Hind III digest.
3. Results
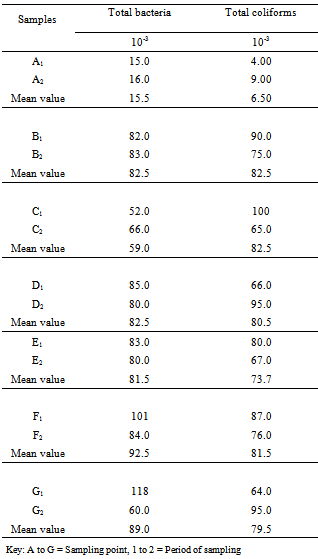
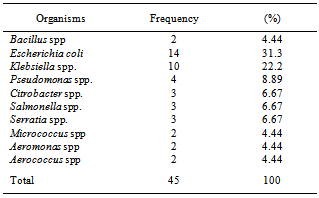
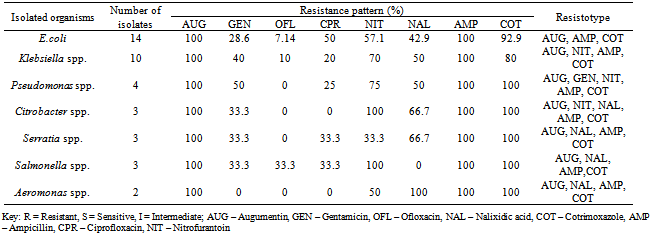
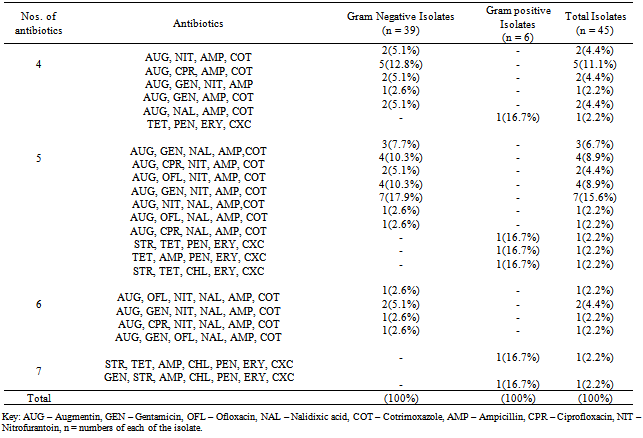
A total of fourteen water samples were collected from seven different points at Arinta waterfall in Ipole-Iloro Ekiti. The average total bacteria count of the water samples A, B, C, D, E, F and G are valued as 5.5 x 103 CFU/ml, 82.5 x 103 CFU/ml, 59.0 x 103 CFU/ml, 82.5 x 103 CFU/ml, 81.5 x 103 CFU/ml, 92.5 x 103 CFU/ml and 89.0 x 103 CFU/ml respectively. While the average total coliform count of the water samples are estimated at 6.5 x 103 CFU/ml, 82.5 x 103 CFU/ml, 82.5 x 103 CFU/ml, 80.5 x 103 CFU/ml, 73.7 x 103 CFU/ml, 81.5 x 103 CFU/ml and 79.5 x 103 CFU/ml respectively. Forty five (45) microbes belonging to ten genera were encountered. Escherichia coli 14(31.1%) had the highest frequency, followed by Klebsiella spp. 10(22.2%), Pseudomonas spp. 4(8.89%), Salmonella spp. Citrobacter spp. and Serratia spp. has 3(6.67%) each while Aeromonas spp. Bacillus spp. Micrococcus spp. and Aerococcus spp. had the least frequency 2(4.44%) each (Table 2). Table 3 shows the antibiotics resistance pattern of the Gram- negative organisms isolated from Arinta water sample using eight antibiotic discs. The isolates demonstrated high level resistance to augumentin, ampicillin, nitrofurantoin and cotrimoxazole while ofloxacin was the most effective of all the antibiotics used. The range of pattern of antibiotics resistance of the organisms isolates were augumentin (100%), amplicillin (100%), cotrimoxazole (80-100%),nitrofurantoin (50-100%), nalixidic acid (0-100%) gentamicin (0-50%), ciprofloxacin (20-50%) and ofloxacin which have the least resistance of (0-33.3%).Table 1. Microbial counts (CFU/ml) of Arinta water samples
 |
| |
|
Table 2. Percentage distribution of microbes from Arinta water samples
 |
| |
|
Table 3. Antibiotics resistance pattern of the Gram-negative organisms from Arinta water samples
 |
| |
|
Table 4. Antibiotics resistance pattern of the Gram-positive organisms isolated
 |
| |
|
Table 5. Phenotype of multiple antibiotic resistance (MAR) patterns of Arinta waterfall sample
 |
| |
|
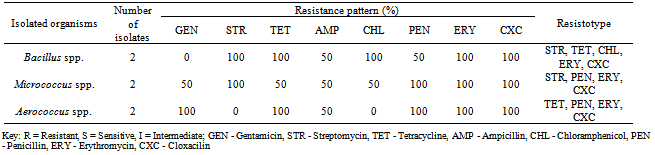
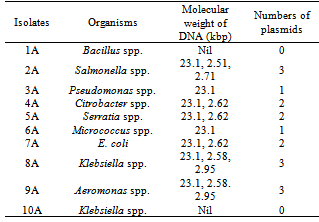
Table 4, shows the antibiotics resistance patterns of isolated Gram-positive organisms. Also the isolates were resistant to Ampicillin (50%) Cloxacillin (100%),Erythromycin (100%), Tetracycline (50-100%) and penicillin (50-100%), while the Gentamicin has 50-100%. Table 5, shows the Multiple Antibiotics Resistance (MAR) patterns of the Forty five (45) bacterial isolates. Multiple antibiotics resistance (MAR) higher amongst gram-negative isolates than gram positive isolates but each gram-positive isolates were resistance to all the antibiotics, cloxacilin, erythromycin and tetracycline. Overall, higher numbers of resistant isolates were identified among Gram-negative isolates. Table 6 shows the plasmid profile patterns and molecular weights of the bacteria isolated from Arinta waterfall. Eight (80%) contained plasmids of which six contained multiple plasmids. Salmonella spp. contained plasmids with molecular weight of 23.1kb, 2.51 kb and 2.71kb (Lane 2A). Klebsiella spp. and Aeromonas spp. carried three plasmids of the same molecular weights of 23.1kb, 2.58kb and 2.95kb (Lanes 8A and 9A). Citrobacter spp., Serratia spp. and E. coli harboured two plasmids each of similar molecular weight of 23.1kb and 2.62kb (Lanes 4A, 5A and 7A). Also Pseudomonas spp. and Micrococcus spp. has 1 plasmid each with molecular weight of 23.1kb (Lanes 3A and 6A), while plasmids were not detected in Bacillus spp. and Klebsiella spp (Lanes 1A and 10A).Table 6. Plasmid profile and molecular weight (Kbp) of isolated bacteria
 |
| |
|
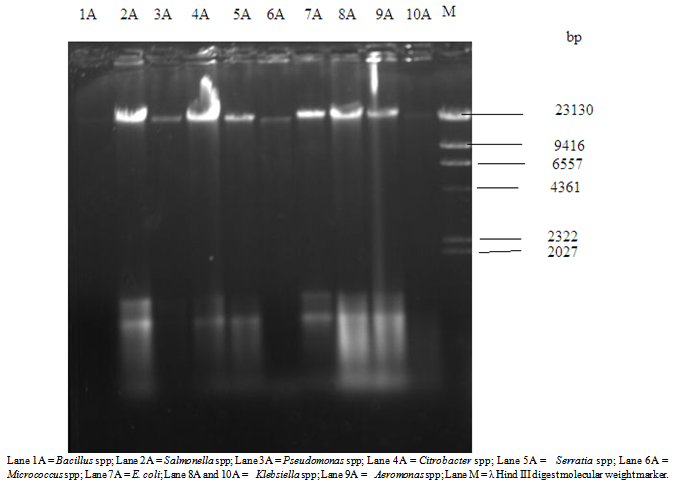 | Figure 2. Plasmid DNA profile of bacterial isolates from Arinta water samples |
4. Discussion
According to guidelines for drinking water quality, the results of the present study indicated that all the water samples tested were of poor microbiological quality. This indicated very high levels of contamination of the Arinta waterfall as well as high diversity of bacterial organisms and high level of antibiotics resistant by isolated microbes. Arinta waterfall is contaminated and threatened by the environmental pollution traced to human activities using the waterfall as tourist and relaxation centre. The presence of such bacteria might result in diseases and poor health of the people using or consuming the water. Although most strains of E. coli are not pathogenic, their presence is an indication of possible presence of pathogenic organisms. Water is considered safe when it is free of Escherichia coli according to[13].The antibiotics susceptibility of the microbes showed higher degree of resistance existing among the aquatic organisms. This is in agreement with the report of[14], who studied Vibrio cholerae during an outbreak of cholera in Lagos in 1997 and reported the isolates were resistant to tetracycline and gentamicin. This study described lower antibiotic resistant to ciprofloxacin and ofloxacin. The highest resistance recorded among the antibiotics was ampicillin, augmentin, tetracycline. These resistance rates are quite high compared to those previously described among organisms isolated from river water where resistance to amikacin was less than 10% and resistance to gentamicin was less than 25%. In a recent study in Poland, high resistance to erythromycin was observed among the gram positive organisms isolated from surface water reaching resistance level above 60%[15]. Increase of antibiotics resistance has been observed among organisms isolated from water sample in Brazil were high indices of resistance to ampicillin and tetracycline were observed[16].[17] also reported high level of resistance in Gram-negative bacteria isolates from rivers in the United States and also revealed that the isolates exhibited multiple resistance to a number of commonly used antibiotics such as ampicillin, tetracycline and penicillin. In this study, ten multi drug resistant bacterial isolates from Arinta waterfall were analyzed for plasmid and this consist of eight (8) Gram-negative organisms and two (2) Gram-positive bacteria. Eight out of ten isolates harboured plasmid out of which six contained more than one plasmid. All of the eight plasmid containing isolates were resistant to augmentin, ampicillin and tetracycline. Meanwhile, plasmid analysis as one of the way to know whether gene coding for antibiotic resistance is located on the plasmid DNA or chromosome DNA. Different plasmids co-existed in the same host cell. This agrees with the finding of[18] who isolated forty five antibiotic resistant bacteria from waste water samples and detected thirty one (31) plasmids in fourteen (14) antibiotic resistant strains with ten (10) carrying multiple plasmids. The demonstration of high-molecular weight plasmids in the isolates and the expression of multidrug resistance (MDR) were plasmid -medicated.[19] showed that several enteric bacterial strains possessing both antibiotic resistance and high molecular weight plasmid could transfer their resistance to other recipient bacteria. The transmissibility of resistance (R) genes and plasmids poses public health risk, considering the vast potential of host presented by microbial populations in the water environment.[17] had in a study of antibiotic resistance of gram-negative bacteria in rivers in the united demonstrated that resistance to ampicillin and other antibiotics drugs (including ciprofloxacin, tetracycline, chloramphenicol, streptomycin and cotrimoxazole) was plasmid mediated. The occurrence of plasmid mediated multi-drug resistant in bacteria in these surface waters heightens the public health. Therefore, the result of this study shows that the plasmid harboured by eight (8) isolates out of the ten isolates might be acquired. Pathogenic bacteria from the water samples were found to be resistance to gentamicin, augumentin, ampicillin, tetracycline and other commonly used antibiotics. Multidrug resistance and plasmid were observed in the bacterial isolates. The occurrence of plasmid mediated multidrug resistant in bacteria in this surface water lightens the public health concern. It can be concluded that this study showed a need for continuous pollutions monitoring programme of the surface water in Nigeria.
References
| [1] | William, C., Sonzogoni, P., Standridge, J., Bussen, M. (2002). Madison preservation and survival of E. coli in well water samples Wisconsin state laboratory of Hygiene, University of Wisconsin. Submitted for routine analysis. |
| [2] | Borchard, M. A., Naas, N. L and Hunt, R. J. (2004). Vulnerability of drinking water wells in La crose, Wisconsin to Enteric-virus contamination from surface water contributions. J. Enteric viral Dis., 2: 12 – 27. |
| [3] | Admassu, M.M., Wubshet, M and Gelaw, B. (2004). A survey of Bacteriological quality of drinking water in north Gondar, Ethiopian Journal of Health Development, 18 (2) 112-115. |
| [4] | World Health Organization (1984). International Standard for drinking water. Expert committee of inter-national standard for drinking water.Geneva 32; Swit- zerland. |
| [5] | Odeyemi, A.T and Agunbiade, R.O. (2012). Bacterio logical and metal analyses of water samples from Awotunde fish pond and river. Scientific Journal of Microbiology.1 (2): 48-54. |
| [6] | Baquero, F., Martinez, J.L and Canton, R. (2008). Antibiotics and antibiotic resistance in water environments. Curr. Opin. Biotechnol., 19:260–265. |
| [7] | Martinez, J. L. (2008). Antibiotics and antibiotic resistance genes in natural environments. Science, 321:365–367. |
| [8] | Berry, D., Xi, C and L. Raskin. (2006). Microbial ecology of drinking water distribution systems. Curr. Opin. Biotechnol., 17:297–302. |
| [9] | American Public Health Association (APHA) (1995). Standard methods for the examination of water and wastewater. 19th ed. American Public Health Association, Washington, D.C. |
| [10] | Barrow, G.I and Feltham, R.K.A. (1993). Cowan and Steel's Manual for the Identification of Medical Bacteria, 3rd edn. Cambridge: Cambridge University Press. |
| [11] | Bauer, A. W., Kirby, W. M., Sherris, J. C and Turck, M. (1966). Antibiotic susceptibility testing by a standar dized single disc method. Am. J. Clin. Pathol, 45. 493 – 496. |
| [12] | Birnboim, H.C and Dolly, J. (1979). A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic acids Research 7: 1513-1523. |
| [13] | Wanke, C. A., Croven, S and Goss, C. (1990). Characterization of binding Escherichia coli strains which are enteropathogens to small bowel mucin. In-fect.immun., 58; 794 – 800. |
| [14] | Idika, N. (1999). The efficacy of water purification methods in the control of Diarrhoeal pathogen in rural communities of Lagos state; Ph.D. Thesis: University of Lagos: Nigeria. |
| [15] | Luczkiewicz, A., Jankowska, K., Kurlenda, J and Olanczuk - Neyman, K. (2010). Identification and anti-Microbial resistance of Enterococcus spp. isolated from surface water. Water Sci. Technol., 62(2): 466 – 473. |
| [16] | Viera, R. H., Carvoiho, E. M., Carvolho, F. C., Silva, C. M., Sousa, O. V and Rodrigues, D. P. (2010). Antimicrobial susceptibility of Escherichia coli isolated from shrimp (litopenaeus vannamel and pond environment in North eastern Brazil. J. Environ. Sci. Health B., 45 (3): 198 – 203. |
| [17] | Ash, R. J., Mauck, B and Morgan, M. (2002). Antibiotic resistance of gram – negative bacteria in rivers United States. Emerg – infect. Dis., 8 :713 – 716. |
| [18] | Fujita, M., Ike, M and Suzuki, H. (1993). Screening of Plasmid from wastewater bacteria. Water Res., 27, 949– 953. |
| [19] | Mcpherson, P and Gealt, M. (1986). Isolation of indigenous wastewater bacterial strains capable of mobilizing plasmid PBR 325. Appl. Environ. Microbial., 51, 904 – 909. |



 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML




