-
Paper Information
- Next Paper
- Previous Paper
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
Journal of Microbiology Research
p-ISSN: 2166-5885 e-ISSN: 2166-5931
2012; 2(4): 95-102
doi: 10.5923/j.microbiology.20120204.06
Detection of Legionella spp. in Natural and Man-made Water Systems Using Standard Guidelines
Borges A. 1, 2, Simões M. 2, Martínez-Murcia A. 3, Saavedra M. J. 1
1Department of Veterinary Science - CECAV, University of Trás-os-Montes and Alto Douro, Vila Real, 5000-801, Portugal
2Department of Chemical Engineering - LEPAE, Faculty of Engineering, University of Porto, Porto, 4200-465, Portugal
3Molecular Diagnostics Center, Biomolecular Technologies S.L., Alicante, Spain
Correspondence to: Simões M. , Department of Chemical Engineering - LEPAE, Faculty of Engineering, University of Porto, Porto, 4200-465, Portugal.
| Email: |  |
Copyright © 2012 Scientific & Academic Publishing. All Rights Reserved.
Infections caused by Legionella spp. are considered at the present time, an emerging public health problem and are linked to high rates of mortality and morbidity, if not properly treated. In this study were analyzed 54 samples of water from 8 counties at Northern Portugal, with the aim of obtaining a collection of strains of the genus Legionella and to characterize them genetically and phenotypically. Another objective of this study was to evaluate the effectiveness of the technique of cultivation, a standard method according to International Organization for Standardization ISO 11731:1998, for detection and enumeration of species of Legionella. For laboratory processing, after the filtration of samples (1 L), the filtrate was resuspended in sterile distilled water (5 ml). Heat treatment for selective inhibition of non-Legionella bacteria was performed. Subsequently, 100 μl of the suspension was spread in GVPC selective agar medium, and incubated (7 to 10 days) at 37 ℃. Colonies that were morphologically characteristic of the genus were sub-cultured onto BCYE agar and blood agar for verification. According to the procedure recommended by the standard method, only the colonies which grew in BCYE agar and not on blood agar were considered as suspected Legionella strains. The identification of these initially selected colonies was performed by sequencing the 16S rRNA gene, which revealed that none of the isolates were identified as belonging to the genus Legionella. However, through the ISO 11731:1998 they were interpreted as positive, corresponding therefore to “false-positive” results. The methods used in this study allowed the isolation of a number of isolates (40), which form an independent group of all genus of the family Chitinophagaceae outlined so far, and that by their phylogenetic distance might be a genus not yet described and therefore a new species. The results obtained, highlighted the importance of using culture and genetic methods in parallel for the proper identification of microorganisms.
Keywords: Water, Legionella, ISO 11731:1998, False positives, 16S rRNA gene sequencing
Article Outline
1. Introduction
- Legionella is an organism of public health interest due to its ability to cause infection in susceptible humans and it’s near ubiquitous presence in heated water systems[1]. The genus Legionella comprises 71 distinct serogroups from more than 50 known species[2-4] and new species are frequently described[5-7]. Legionella pneumophila, was first recognized in 1977 following an epidemic of acute pneumonia in Philadelphia[8], is the etiological agent of the majority of cases of legionellosis, and the best part of cases have been attributed to L. pneumophila serogroup 1[3,9]. In addition, at least 21 other species of Legionella have been related with human infections[2,10]. These organisms may be a cause of nosocomial and community-acquired infections, principally in immunocompromised patients[10-12]. Legionellosis outbreaks are often associated with highmortality rates[13].Bacteria have been found in watery soils, natural orman-made aquatic environments, such as rivers, lakes, ponds, mud, and cooling towers or water distribution systems[2,14-17]. Legionella bacteria do exist as free-livingplanktonic forms in the environment, intracellular parasites of protozoans, and/or inhabitants of mixed community biofilms[1,18]. Thus, the diversity of types and the ubiquitous occurrence of legionellae in water environments or moist soil make it difficult to identify epidemic strains, and outbreaks of legionellosis have been associated mainly with contamination of man-made aquatic environments[19].Legionnaires' disease can be acquired by the inhalation of contaminated droplet aerosols or by microaspiration of contaminated water[10,15]. However transmission of Legionella from person to person has never been observed, and the prevention needs to focus on the elimination of this pathogen from water and systems that produce aerosol. Thus, rapid and accurate detection of Legionella in water systems is extremely importante for hazard prediction and the elimination of Legionella from potential infection sources[20]. The detection of water contaminated by Legionella has been usually performed using culture-based methods, approved by the International Organization for Standardization[21] and the French organization for standardization[22]. Culture is important for identifying and typing Legionella strains during epidemics but have several limitations, namely long incubation times. This problem makes culture inappropriate for preventive actions and rapid response in emergency situations. Another weakness is the occurrence of viable but nonculturable (VBNC) bacteria. Furthermore, the sensitivity of Legionella detection based on culture methods depends largely on the physiological state of the cells[20,23].Therefore, it is important to improve the current testing for Legionella and develop rapid and accurate tests. In recent years, different PCR-based methods (based on 16S rRNA, 5S rRNA and 23S rRNA genes and on the mip gene encoding the macrophage infectivity potentiator gene of L. pneumophila) for detection and quantification of Legionella in water samples have been described and can moderate the main drawbacks of culture-based methods[24-28]. The development of more rapid, culture-independent methods able to discriminating between live and dead cells is very important for assessing Legionella infection risks and preventing legionellosis[23].The aim of the present study was to characterize presumptive bacteria from the genus Legionella isolated from water samples from various sources, and also to evaluate the efficiency of detection of Legionella species by the technique of culture, according to the recommendations of the International Standard ISO 11731:1998.
2. Materials and Methods
- Sample collectionA total of 54 water samples were collected at different natural aquatic environmental sources (fountains, springs, rivers, wells and lakes), and from man-made systems (decorative fountains, tap water, water cylinder and the water pipe condensation of the cooling towers of air conditioning) of eight counties from Northern Portugal (Table 1), for a period of 10 months (January and October of 2008). Water samples were collected in sterile 1000 ml polyethylene bottles and samples were delivered to the laboratory within one day.Isolation of legionellae from water samplesIsolation of Legionella from water samples was performed by culture according to the recommendations of the International Standard method ISO 11731:1998 (Water quality - Detection and enumeration of Legionella)[21], based on filtration procedure and culture of bacteria on selective media. Samples of 1000 ml of water were concentrated by filtration on a 0.2 µm pore-diameter polycarbonate membrane (Isopore, Millipore, Ireland). After filtration, bacteria collected on the membranes were resuspended in 5 ml of water and shaken vigorously. Heat treatment for selective inhibition of non-Legionella bacteria were performed as described previously[21,24,29,30]. Briefly, 1 ml of the concentrate was treated at 50 ± 1℃ for 30 ± 1 min. Subsequently 0.1 ml of the suspension was spread on a 90-mm Petri dish containing BCYE agar supplemented with glycine, vancomycin, polymyxin B and cycloheximide (GVPC selective agar medium) (Becton, Dickinson and Company, USA). The inoculated plates were then incubated for 7 to 10 days at 37±1℃ under aerobic conditions and humidified atmosphere. Colonies were counted after 3, 5, and 10 days. Smooth colonies showing a yellowish or sometimes a yellow-green or grayish-white color were counted as suspicious legionellae to be confirmed. Up to 5 to 7 colonies of suspected Legionella were subcultured onto BCYE agar (without antibiotics) (Becton, Dickinson and Company, USA), and blood agar (alternatively we can use BCYE agar without L-cysteine) for confirmation. The isolated colonies growing only on BCYE agar but not on blood agar were considered to be Legionella colonies. No further confirmatory tests, namely direct or indirect immunofluorescence and latex agglutination, for cysteine-dependent colonies, were carried out. A positive control with L. pneumophila CECT 7109 was used.
|
3. Results
- Strains isolated during this work were obtained from 54 water samples, collected from 8 counties of Northern Portugal and 9 different sampling sites (fountains, springs, wells, rivers, lakes, recreational sources, tap water, cylinders and air conditioning) (Table 1). From a total of 128 isolated bacteria, 49 grew only on BCYE agar and the remaining 79 grew in both culture media (BCYE agar and blood agar). The first group was then considered presumptive Legionella species, in accordance with ISO 11731:1998. Biochemical tests were carried out in two isolates (VR17a4 and VR17a5) of this group. All results were the same for the two isolates. The tests for oxidase, catalase and hippurate hydrolysis were positive. However, tests for gelatinase, urease, nitrate reduction and utilization of glucose and other carbohydrates were negative. The suspected colonies (49) were then directly identified by molecular methods, performed by sequencing the 16S rRNA gene (Table 3). Gene sequencing results revealed that none of the isolates were identified as belonging to the genus Legionella. However, through the indications of the ISO 11731:1998 they were interpreted as positive, corresponding therefore to “false-positive” results. This occurrence emphasizes the importance of the use of methods at the molecular level, for unambiguous identification of strains. In Table 3 are shown the results of the identification of presumptive Legionella species, by 16S rRNA gene sequencing. The isolate F1L was obtained from a water sample of a fountain, and identified as Bacillus pumilus. The isolates VR26a1, VR27a1 and VR27a1F were identified as Burkholderia fungorum. The identification of the bacteria VR27a3 provided two different species, Sphingomonas melonis and Sphingomonas aquatilis. Those species belong to the same bacterial genus, based on the partial sequence.
|
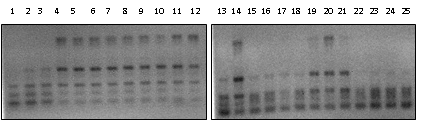 | Figure 1. RAPD image with the primer ERIC (ERIC-1R and ERIC-2). The numbers in the image correspond to the isolates: 1 - VR17a2, 2 - VR17a3, 3 - VR17a4, 4 - VR17a5, 5 - VR25a1, 6 - VR25b1, 7 - VR25a2, 8 - VR25a4, 9 - VR25a5, 10 - VR25a6, 11 - VR25a7, 12 - VR25a8, 13 - VR25b2, 14 - VR25b3, 15 - VR25b4, 16 - VR25b5, 17 - VR25c2, 18 - VR25d2, 19 - VR25e2, 20 - VR25a1F, 21 - VR25a2F, 22 - VR25a3F, 23 - VR25a4F, 24 - VR27a2, 25 - VR27a2F. |
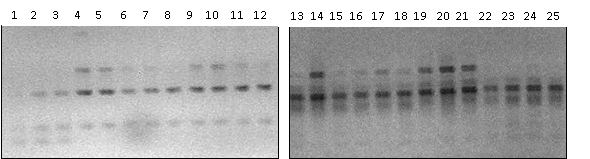 | Figure 2. RAPD image with primers OPA 20 (1 -VR17a2, 2 - VR17a3, 3 - VR17a4, 4 - VR17a5, 5 - VR25a1, 6 - VR25b1, 7 - VR25a2, 8 - VR25a4, 9 - VR25a5, 10 - VR25a6, 11 - VR25a7, 12 - VR25a8) and OPA 16 (13 - VR25b2, 14 - VR25b3, 15 - VR25b4, 16 - VR25b5, 17 - VR25c2, 18 - VR25d2, 19 - VR25e2, 20 - VR25a1F, 21 - VR25a2F, 22 - VR25a3F, 23 - VR25a4F, 24 - VR27a2, 25 - VR27a2F). |
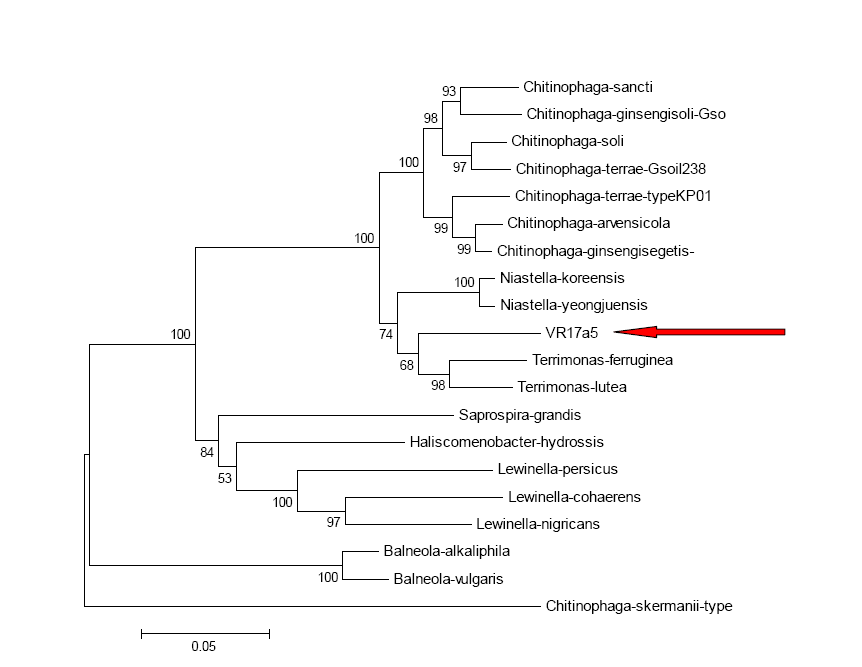 | Figure 3. Phylogenetic tree based on the sequence of the 16S rRNA, showing the phylogenetic position of strain VR17a5 (representative strain) |
4. Discussion
- Legionella species have growth requirement for L-cysteine and without this amino acid, they are unable to grow. Usually colonies with typical morphology on selective media are subcultured onto BCYE and BCYE without L-cysteine (or blood agar). The isolates that grow on BCYE but fail to grow on BCYE without L-cysteine, can be presumptively identified as Legionella spp.[24,47]. Closer identification can be carried out using latex agglutination, direct immunofluoresence or molecular techniques. Some phenotypic tests have lack of sensitivity, and inability for accurately detect all clinically important Legionella species and serogroups, and should not be used as a single tool[48].Using the traditional biochemical tests, bacteria of the genus Legionella cannot be identified with any degree of certainty. Many of the species can only be correctly identified using molecular methods. There is a broad panel of biochemical tests whose usefulness has been demonstrated in identification, but these methods have not been widely used or validated after their initial description[49]. Tests for nitrate reductase and urease are negative for all species. Most strains liquefy gelatin and are reported to be catalase positive and oxidase negative (although they may provide positive results). L. pneumophila and a few other species hydrolyze hippurate[16,19,50]. The legionellae are chemoorganotropic and do not possess a glucose transport system, nor ferment or oxidize other carbohydrates[49,51]. These biochemical characteristics were observed for two selected isolates (VR17a4 and VR17a5). The results obtained are a further indication, in addition to the L-cysteine growth dependence, that these isolates may belong to the genus Legionella.In this study, no additional confirmatory tests, for cysteine-dependent colonies, were performed and the presumptive Legionella spp. were identified by 16S rRNA gene sequencing. The positive results of the cultivation method were not confirmed by genetic identification. At this context, the results obtained can create a problem of economic nature, since, one of the measures to be implemented, after the detection of Legionella, would be the cleaning and disinfection of all contaminated systems, in order to avoid potential cases of infection. It should be noted that sometimes the opposite occurs, i.e. false negatives are obtained, since the technique of cultivation does not detect the presence of Legionella and genetically this microorganism is detected[48,52].Legionella species have been isolated from a wide variety of water types, such as potable water of hospital, industries and hotels, ground and surface water and biofilms[53]. Moreover, several authors[1,3,50,54] have described the isolation of Legionella spp. from showers, cooling towers and boilers. In contrast with those reports, in this work Legionella spp. were not detected in the aquatic environments studied.Despite advances in medium formulations and pretreatment techniques, recovery of Legionella species from water samples can still be quite low, difficult and time consuming[48,52]. They are generally present at very low or undetectable concentrations in freshwater and moist natural aquatic environments. Thus, when working with environmental samples, it is usually necessary to use a concentration technique (centrifugation and/or filtration) of the microflora. However, for clinical or environmental samples it is necessary to eliminate or suppress competitive flora during primary culture. To reduce the growth of unwanted bacteria, the samples can be subjected to a heat treatment (50 ℃ for 30 min) or acid (pH 2.2 for 5 min). This treatment of the samples is performed according to the sampling criteria specified by ISO 11731:1998[21] and was already used in several studies[24,29,51].The water temperature is assumed as the prime factor affecting their incidence[55,56]. Legionellae appear to have a predilection for the warm water encountered in artificial environments, such as man-made systems. Looking for better recovery results, Sanden et al.[57] observed that preincubation of water samples with free-living amoebae for several days (seven days) increased the recovery of Legionella spp., which was related with proliferation of amoebae in the samples. The same observations were done by Bartie et al.[30], who showed that re-incubation of water samples concentrates with autochthonous amoebae improved the culturability of legionellae in a selective medium. Although these approaches are quite selective, due to the improvement of limit of detection or interference with other bacteria, these methods can be time expensive. Culturing is generally accepted as the “golden standard” for Legionella detection in the environment, but the lack of standardization of culturing methods by some laboratories, especially for environmental legionellae, complicates the interpretation of results[58]. This method allows the isolation and the quantification of cultivable Legionella species from environmental and clinic samples but it does have limitations[13,59]. The cultivation method has also the disadvantage of being a very time-consuming technique, requiring selective media and several days of incubation (7 to 10 days) to obtain results[25,59]. In addition, although there is an international consensus standard for culture detection and enumeration of Legionella in water (ISO 11731:1998), the overgrowth of other accompanying bacteria, the presence of viable but non-culturable (VBNC) cells, loss of viability of bacteria after collection and during the sample treatment (concentration stage followed by decontamination with heat or acid), reduced recoveries by the use of antibiotics in the medium and low concentration of legionellae in the samples limit the use of this method[16,24,52,59,60].Bacteria exposed to potentially lethal environmental conditions including nutrient restriction, oxidative stress, heat, UV irradiation, osmotic stress, or sublethal concentrations of antibacterial compounds undergo physiological or morphological alterations that complicate the detection and accurate enumeration of such stressed bacteria using available culture methods[24,58]. We can conclude, for all these reasons, that the populations of Legionella are underestimate by culture methods.There is even the occurrence of “false-positives”, as found in this work, which highlights the importance of using faster genetic methods for the proper identification of microorganisms. Thus, there is a need for improved detection methods.PCR methods, such as quantitative real-time PCR (qPCR), are an alternative tool to the conventional culture method for the detection of slow-growing and fastidious bacteria such as Legionella species[61-63]. Amplification of Legionella DNA by PCR can provide results within a short time. It also has the potential to detect infections caused by any Legionella species and serogroups, in addition to detect non-culturable legionella and allow the manipulation of a large number of samples[3,9,59,64]. The detection of other species besides L. pneumophila is relevant to the diagnosis of Legionnaires' disease, since this disease can be caused by other species, such as L. longbeachae and L. bozemanii. The correct identification of species is also important for epidemiological studies and identification of sources of infection[65]. However, the major disadvantage of PCR it is the inability to evaluate the viability, in other words, by the PCR technique it is not possible to distinguish between viable and nonviable microorganisms, detecting only their presence or absence, while only viable bacteria are able to cause infections in human and represent an interest for public health[20,23,47]. The qPCR is an important complement, but not a substitute, for the standard culture-based methods[66].The development of new and rapid assays that combine both specific detection and viability criteria is essential for monitoring water quality and legionellosis prevention[3,24]. A rapid, sensitive, and specific method for the detection of Legionella is clearly important to identify infected individuals and to expedite cleanup of contaminated water systems (reduction in disinfecting treatment cost) in order to prevent additional cases of infection[25,47].
5. Conclusions
- In conclusion, elucidation of the properties of the Legionella species is needed to assess their potential public health significance and explain the conditions favoring their growth in aquatic environments. The detection tools for Legionella species have been commonly based on culture method investigations. However, this method is not conveniently sensitive and is time-consuming (most Legionella spp. colonies being detected within 7 days). From this study, we can conclude that the utilization of the technique of culture, according to the International Standard ISO 11731:1998, it is possible to have false positives results in the detection of presumptive Legionella species. Therefore, no Legionella spp. were detected in water samples analyzed during this work. Prospective studies using the culture method and molecular methods should be performed in parallel, to provide better and more comprehensive approach to detection of Legionella species. The method used in this study, allowed obtaining a number of isolates, which form an independent group of all genus of the family Chitinophagaceae described so far, and that by their phylogenetic distance may be a genus and therefore a species not yet described.
ACKNOWLEDGEMENTS
- This work was supported by the Operational Programme for Competitiveness Factors – COMPETE and by FCT – Portuguese Foundation for Science and Technology through Project Bioresist - PTDC/EBB-EBI/105085/2008 and the PhD grant awarded to Anabela Borges(SFRH/BD/63398/2009).
References
| [1] | Taylor, M., Ross, K., Bentham, R. 2009. Legionella, protozoa, and biofilms: Interactions within complex microbial systems. Microb Ecol 58:538-547. |
| [2] | Kuroki, H., Miyamoto, H., Fukuda, K., Iihara, H., Kawamura, Y., Ogawa, M., Wang, Y., Ezaki, T., Taniguchi, H. 2007. Legionella impletisoli sp. nov. and Legionella yabuuchiae sp. nov., isolated from soils contaminated with industrial wastes in Japan. Syst Appl Microbiol 30:273-279. |
| [3] | Diederen, B.M.W., Kluytmans, J.A.J.W.,Vandenbroucke-Grauls, C.M., Peeters, M.F. 2008. Utility of real-time PCR for diagnosis of legionnaires' disease in routine clinical practice. J Clin Microbiol 46:671-677. |
| [4] | Tronel, H., Hartemann, P. 2009. Overview of diagnostic and detection methods for legionellosis and Legionella spp. Lett Appl Microbiol 48:653-656. |
| [5] | Park, M.Y., Ko, K.S., Lee, H.K., Park, M.S., Kook, Y.H. 2003. Legionella busanensis sp. nov., isolated from cooling tower water in Korea. Int J Syst Evol Microbiol 53:77-80. |
| [6] | La Scola, B., Birtles, R.J., Greub, G., Harrison, T.J., Ratcliff, R.M., Raoult, D. 2004. Legionella drancourtii sp. nov., a strictly intracellular amoebal pathogen. Int J Syst Evol Microbiol 54:699-703. |
| [7] | Pourcel, C., Visca, P., Afshar, B., D'Arezzo, S., Vergnaud, G., Fry, N.K. 2007. Identification of variable-number tandem-repeat (VNTR) sequences in Legionella pneumophila and development of an optimized multiple-locus VNTR analysis typing scheme. J Clin Microbiol 45:1190-1199. |
| [8] | Fraser, D.W., Tsai, T.R., Orenstein, W. 1977. Legionnaires' disease. Description of an epidemic of pneumonia. N Engl J Med 297:1189-1197. |
| [9] | Jarraud, S., Reyrolle, M., Meugnier, H., Forey, F., Etienne, J. 2007. Legionnaires disease. Presse Med 36:279-287. |
| [10] | Kümpers, P., Tiede, A., Kirschner, P., Girke, J., Ganser, A., Peest, D. 2008. Legionnaires' disease inimmunocompromised patients: A case report of Legionella longbeachae pneumonia and review of the literature. J Med Microbiol 57:384-387. |
| [11] | Stojek, N.M., Szymańska, J., Dutkiewicz, J. 2008. Gram-negative bacteria in water distribution systems of hospitals. Ann Agric Environ Med 15:135-142. |
| [12] | Liguori, G., Bagattini, M., Gallè, F., Quartucci, V., Di Onofrio, V., Negrone, M., Triassi, M. 2009. Microbiological evaluation of the efficacy of two new biodetergents on multidrug-resistant nosocomial pathogens. Ann Clin Microbiol Antimicrob 8:1-6. |
| [13] | Yaradou, D.F., Hallier-Soulier, S., Moreau, S., Poty, F., Hillion, Y., Reyrolle, M., André, J., Festoc, G., Delabre, K., Vandenesch, F., Etienne, J., Jarraud, S. 2007. Integrated real-time PCR for detection and monitoring of Legionella pneumophila in water systems. Appl Environ Microbiol 73:1452-1456. |
| [14] | Alp, E., Voss, A. 2006. Ventilator associated pneumonia and infection control. Ann Clin Microbiol Antimicrob 5:1-11. |
| [15] | Diederen, B.M.W. 2008. Legionella spp. and Legionnaires' disease. J Infect 56:1-12. |
| [16] | Bartram, J., Chartier, Y., Lee, J.V., Pond, K., Surman-Lee, S. 2007. Legionella and the prevention of legionellosis. World Health Organization (WHO). Geneva. SBN: 9241562978. |
| [17] | Fields, B.S., Ed. 2008. Legionella in the environment. In: Legionella pneumophila: pathogenesis and immunity. Hoffman, P., Friedman, H., Bendinell, M. (Ed.), Springer. |
| [18] | Declerck, P., Behets, J., Van Hoef, V., Ollevier, F. 2007. Replication of Legionella pneumophila in floating biofilms. Curr Microbiol 55:435-440. |
| [19] | Fields, B.S., Benson, R.F., Besser, R.E. 2002. Legionella and legionnaires' disease: 25 Years of investigation. Clin Microbiol Rev 15:506-526. |
| [20] | Mérault, N., Rusniok, C., Jarraud, S., Gomez-Valero, L., Cazalet, C., Marin, M., Brachet, E., Aegerter, P., Gaillard, J.L., Etienne, J., Herrmann, J.L., Lawrence, C., Buchrieser, C. 2011. Specific real-time PCR for simultaneous detection and identification of Legionella pneumophila serogroup 1 in water and clinical samples. Appl Environ Microbiol 77:1708-1717. |
| [21] | International Standards Organisation. 1998. Water quality - detection and enumeration of Legionella. International standard ISO 11731. International Standards Organisation (International Organization for Standardization), Geneva, Switzerland. |
| [22] | Association Franc¸aise de Normalisation. 2003. Water quality. Detection and enumeration of Legionella spp. and Legionella pneumophila. Method by direct inoculation and after concentration by membrane filtration of centrifugation. AFNOR NF T90-431. Association Française de Normalisation, La Plaine Saint-Denis, France. |
| [23] | Delgado-Viscogliosi, P., Solignac, L., Delattre, J.M. 2009. Viability PCR, a culture-independent method for rapid and selective quantification of viable Legionella pneumophila cells in environmental water samples. Appl Environ Microbiol 75:3502-3512. |
| [24] | Delgado-Viscogliosi, P., Simonart, T., Parent, V., Marchand, G., Dobbelaere, M., Pierlot, E., Pierzo, V., Menard-Szczebara, F., Gaudard-Ferveur, E., Delabre, K., Delattre, J.M. 2005. Rapid method for enumeration of viable Legionella pneumophila and other Legionella spp. in water. Appl Environ Microbiol 71:4086-4096. |
| [25] | Nazarian, E.J., Bopp, D.J., Saylors, A., Limberger, R.J., Musser, K.A. 2008. Design and implementation of a protocol for the detection of Legionella in clinical and environmental samples. Diagn Microbiol Infect Dis 62:125-132. |
| [26] | Wellinghausen, N., Frost, C., Marre, R. 2001. Detection of Legionellae in Hospital Water Samples by Quantitative Real-Time LightCycler PCR. Appl Environ Microbiol 67:3985-3993. |
| [27] | Villari, P., Motti, E., Farullo, C., Torre, I. 1998. Comparison of conventional culture and PCR methods for the detection of Legionella pneumophila in water. Lett Appl Microbiol 27:106-110. |
| [28] | Van der Zee, A., Verbakel, H., De Jong, C., Pot, R., Bergmans, A., Peeters, M., Schneeberger, P., Schellekens, J. 2002. Novel PCR-probe assay for detection of and discrimination between Legionella pneumophila and other Legionella species in clinical samples. J Clin Microbiol 40:1124-1125. |
| [29] | Lück, P.C., Helbig, J.H., Schuppler, M. 2002. Epidemiology and laboratory diagnosis of Legionella infections. J Lab Med 26:174-182. |
| [30] | Bartie, C., Venter, S.N., Nel, L.H. 2003. Identification methods for Legionella from environmental samples. Water Res 37:1362-1370. |
| [31] | Benson, R.F., Thacker, W.L., Daneshvar, M.I., Brenner, D.J. 1996. Legionella waltersii sp. nov. and an unnamed Legionella genomospecies isolated from water in Australia. Int J Syst Bacteriol 46:631-634. |
| [32] | Fox, K.F., Brown, A. 1989. Application of numerical systematics to the phenotypic differentiation of legionellae. J Clin Microbiol 27:1952-1955. |
| [33] | Hebert, G.A. 1981. Hippurate hydrolysis by Legionella pneumophila. J Clin Microbiol 13:240-242. |
| [34] | Orrison, L.H., Cherry, W.B., Fliermans, C.B. 1981. Characteristics of environmental isolates of Legionella pneumophila. Appl Environ Microbiol 42:109-115. |
| [35] | Pine, L., Hoffman, P.S., Malcolm, G.B. 1984. Whole-cell peroxidase test for identification of Legionella pneumophila. J Clin Microbiol 19:286-290. |
| [36] | Weaver, R.E., Feeley, J.C., (Eds.), 1979. Cultural and biochemical characterization of Legionnaires’ disease bacterium, in: G.L. Jones, G.A. Herbert (Eds.), Legionnaires’, the Disease, the Bacterium, and Methodology, Centers for Disease Control, Atlanta, GA, pp. 20-25. |
| [37] | Martinez-Murcia, A.J., Benlloch, S., Collins, M.D. 1992. Phylogenetic interrelationships of members of the genera Aeromonas and Plesiomonas as determined by 16S ribosomal DNA sequencing: Lack of congruence with results of DNA-DNA hybridizations. Int J Syst Bacteriol 42:412-421. |
| [38] | Thompson, J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F., Higgins, D.G. 1997. The CLUSTAL X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876-4882. |
| [39] | Martínez-Murcia, A.J. 1999. Phylogenetic positions of Aeromonas encheleia, Aeromonas popoffii, Aeromonas DNA hybridization Group 11 and Aeromonas Group 501. Int J Syst Bacteriol 49:1403-1408. |
| [40] | Saavedra, M.J., Figueras, M.J., Martínez-Murcia, A.J. 2006. Updated phylogeny of the genus Aeromonas. Int J Syst Evol Microbiol 56:2481-2487. |
| [41] | Saavedra, M.J., Perea, V., Fontes, M.C., Martins, C., Martínez-Murcia, A. 2007. Phylogenetic identification of Aeromonas strains isolated from carcasses of pig as new members of the species Aeromonas allosaccharophila. Antonie van Leeuwenhoek 91:159-167. |
| [42] | Kimura, M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111-120. |
| [43] | Saitou, N., Nei, M. 1987. The Neibhbour-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406-425. |
| [44] | Kumar, S., Tamura, K., Jakobsen, I.B., Nei, M. 2001. MEGA2: Molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245. |
| [45] | Nkongolo, K.K., Nsapato, L. 2003. Genetic diversity in Sorghum bicolor (L.) Moench accessions from different ecogeographical regions in Malawi assessed with RAPDS. Genet Resour Crop Evol 50:149-156. |
| [46] | Davin-Regli, A., Bollet, C., Chamorey, E., Colonna D'Istria, V., Cremieux, A. 1998. A cluster of cases of infections due to Aeromonas hydrophila revealed by combined RAPD and ERIC-PCR. J Med Microbiol 47:499-504. |
| [47] | Cooper, I.R., Meikle, S.T., Standen, G., Hanlon, G.W., Santin, M. 2009. The rapid and specific real-time detection of Legionella pneumophila in water samples using Optical Waveguide Lightmode Spectroscopy. J Microbiol Methods 78:40-44. |
| [48] | Murdoch, D.R. 2003. Diagnosis of Legionella infection. Clin Infect Dis 36:64-69. |
| [49] | Edelstein, P.H., Cianciotto, N.P. 2006. Legionella Species and Legionnaires’ Disease. Prokaryotes 6:988-1033. |
| [50] | Muder, R.R., Yu, V.L. 2002. Infection due to Legionella species other than L. pneumophila. Clin Infect Dis 35:990-998. |
| [51] | Murray, P.R., Rosenthal, K.S., Pfaller, M.A., (Eds.), 2005. Legionella. In: Medical Microbiology, 5th edn. Elsevier Science, pp. 391-340. |
| [52] | Blanco, S., Prat, C., Sánchez, M.D., Ferrer, D., Pellicer, T., Haba, L., Latorre, I., Vilaplana, C., Ausina, V., Domínguez, J. 2008. Evaluation of a Legionella urinary antigen enzyme immunoassay for rapid detection of Legionella pneumophila in water samples. Int J Hyg Environ Health 211:168-171. |
| [53] | Carvalho, F.R.S., Foronda, A.S., Pellizari, V.H. 2007. Detection of Legionella pneumophila in water and biofilm samples by culture and molecular methods from man-made systems in São Paulo - Brazil. Brazil J Microbiol 38:743-751. |
| [54] | Delaedt, Y., Daneels, A., Declerck, P., Behets, J., Ryckeboer, J., Peters, E., Ollevier, F. 2008. The impact of electrochemical disinfection on Escherichia coli and Legionella pneumophila in tap water. Microbiol Res 163:192-199. |
| [55] | Lee, J.V., West, A.A. 1991. Survival and growth of Legionella species in the environment. J Appl Bacteriol 121S-129S. |
| [56] | Ozlem Sanli-Yurudu, N., Kimiran-Erdem, A., Cotuk, A. 2007. Studies on the efficacy of Chloramine T trihydrate (N-chloro-p-toluene sulfonamide) against planktonic and sessile populations of different Legionella pneumophila strains. Int J Hyg Environ Health 210:147-153. |
| [57] | Sanden, G.N., Morrill, W.E., Fields, B.S., Breiman, R.F., Barbaree, J.M. 1992. Incubation of water samples containing amoebae improves detection of legionellae by the culture method. Appl Environ Microbiol 58:2001-2004. |
| [58] | Kozioł-Montewka, M., Magryś, A., Stojek, N., Palusińska-Szysz, M., Danielak, M., Wójtowicz, M., Niewiedzioł, J., Koncewicz, R., Niedźwiadek, J., Paluch-Oleś, J., Trzeciak, H., Drozański, W., Dutkiewicz, J. 2008. Monitoring Legionella species in hospital water systems. Link with disease and evaluation of different detection methods. Ann Agric Environ Med 15:143-147. |
| [59] | Diederen, B.M.W., de Jong, C.M.A., Aarts, I., Peeters, M.F., van der Zee, A. 2007. Molecular evidence for the ubiquitous presence of Legionella species in Dutch tap water installations. J Water Health 5:375-383. |
| [60] | Allegra, S., Berger, F., Berthelot, P., Grattard, F., Pozzetto, B., Riffard, S. 2008. Use of flow cytometry to monitor Legionella viability. Appl Environ Microbiol 74:7813-7816. |
| [61] | Dusserre, E., Ginevra, C., Hallier-Soulier, S., Vandenesch, F., Festoc, G., Etienne, J., Jarraud, S., Molmeret, M. 2008. A PCR-based method for monitoring Legionella pneumophila in water samples detects viable but noncultivable legionellae that can recover their cultivability. Appl Environ Microbiol 74:4817-4824. |
| [62] | Joly, P., Falconnet, P.A., André, J., Weill, N., Reyrolle, M., Vandenesch, F., Maurin, M., Etienne, J., Jarraud, S. 2006. Quantitative real-time Legionella PCR for environmental water samples: Data interpretation. Appl Environ Microbiol 72:2801-2808. |
| [63] | Behets, J., Declerck, P., Delaedt, Y., Creemers, B., Ollevier, F. 2007. Development and evaluation of a Taqman duplex real-time PCR quantification method for reliable enumeration of Legionella pneumophila in water samples. J Microbiol Methods 68:137-144. |
| [64] | Wilson, D.A., Yen-Lieberman, B., Reischl, U., Gordon, S.M., Procop, G.W. 2003. Detection of Legionella pneumophila by real-time PCR for the mip gene. J Clin Microbiol 41:3327-3330. |
| [65] | Bencini, M.A., Van Den Brule, A.J.C., Claas, E.C.J., Hermans, M.H.A., Melchers, W.J.G., Noordhoek, G.T., Salimans, M.M.M., Schirm, J., Vink, C., Van Der Zee, A., Jansen, R. 2007. Multicenter comparison of molecular methods for detection of Legionella spp. in sputum samples. J Clin Microbiol 45:3390-3392. |
| [66] | Morio, F., Corvec, S., Caroff, N., Le Gallou, F., Drugeon, H., Reynaud, A. 2008. Real-time PCR assay for the detection and quantification of Legionella pneumophila in environmental water samples: Utility for daily practice. Int J Hyg Environ Health 211:403-411. |
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-Text HTML
Full-Text HTML