-
Paper Information
- Next Paper
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
Journal of Microbiology Research
p-ISSN: 2166-5885 e-ISSN: 2166-5931
2012; 2(3): 47-50
doi: 10.5923/j.microbiology.20120203.01
Molecular Characterization of Seven Different Species of Aspergillus through Random Amplified Polymorphic DNA (RAPD) and Enzyme Analysis
Saba Irshad , Rabiya Nawab
Institute of Biochemistry and Biotechnology University of the Punjab Lahore, 54590, Pakistan
Correspondence to: Saba Irshad , Institute of Biochemistry and Biotechnology University of the Punjab Lahore, 54590, Pakistan.
| Email: |  |
Copyright © 2012 Scientific & Academic Publishing. All Rights Reserved.
The aim of this study was to characterize seven species of Aspergillus at molecular level, using random amplification of polymorphic DNA (RAPD). RAPD- PCR conditions were optimized for two primers of series B, GL Decamer B-09 and GL Decamer B-10 out of 10 total primers. RAPD results were evaluated by a statistical software Minitab and a phylogenetic tree was prepared. GL Decamer B-09 showed 38 bands and GL Decamer B-10 gave 46 bands, showing 50% and 57% similarity respectively, among species. Biochemical characterization was done by screening of zones production with particular enzyme activity of each specie resulted in particular substrate degradation.
Keywords: Aspergillus, RAPD, Minitab, Pectinase activity, Phylogeny
Article Outline
1. Introduction
- There are an estimated 1.5 million fungal species of which around 70,000 have been described so far[1]. Aspergillus is a filamentous and ubiquitous fungus found in nature and is identified at species level by using the differential culture media. A total of 205 Aspergillus species are reported which includes 153(75%) environmental and 52 (25%) clinical Aspergilli[2]. Aspergillus niger is economically important fermentation organism used for the production of citric acid and it represents highest yield bioprocess, currently in use by industry[3]. Aspergillus fumigatus is considered to be the most frequent isolate from clinically immunocompromised patients, but other important species include A. flavus, A. niger and A. terreus as a cause of opportunistic infections[4]. Phenotypic and genotypic correlations provide strong evidence for differentiation of A. flavus from A. oryzae, though there are very minor phenotypic and genotypic differences[5]. Bacterial strains are effective in reducing soil populations of mycotoxigenic fungi; thereby reduce fungal spore formation and crop plant infection via airborne transmission[6].PCR-based technique, involving the random amplification of polymorphic DNA (RAPD) has been used for assessing genomic variability among a wide range of culture collection strains of Aspergillus and related species[7]. The utility of DNA markers as RAPD-DNA employ it as well established sample molecular marker tool for detecting genetic variability for many phytopathogenic fungi[8].DNA Polymorphisms based on differences in DNA sequences, have advantages over protein polymorphisms[9]. The application of the random amplification of polymorphic DNA (RAPD) assay to the human pathogen A. fumigatus was described in the year 1993[10]. RAPD fingerprinting is used to gain rapid and precise information about genetic similarities and dissimilarities of different Aspergillus species. RAPD fingerprints of A. niger, A. flavus and A. parasiticus revealed polymorphism in 37, 59, 51% of the analyzed Aspergillus species[11].RAPD polymorphism results from a nucleotide base change, an insertion or deletion that alters the primer binding sites. This product can be polymorphic and may be used as genetic markers for extensive genetic variation analysis[12]. One advantage of this technique is that the primers are universal and they can be used for genomic analysis of a wide variety of species[13].The research project of analyzing genetic diversity by RAPD PCR is very useful to detect similarities and differences in different fungal species. Random Amplified Polymorphic DNA, more or less randomly distributed in the whole genome[14]. They span the majority of the chromosome and map both proximal and distal to the centromeres and are also able to map novel chromosomal regions. In the present study we have characterized seven different Aspergillus strains i.e A. niger, A. nidulans, A. parasiticus, A. flavus, A. japonicus, A. oryzae and A. fumigatus. The aim of the present study was to characterize seven different species of fungi at molecular level that leads to further elucidation of genetic diversity.
2. Material and Methods
2.1. Source of Fungi
- The fungal samples were obtained from First Fungal Culture Bank of University of the Punjab Lahore at Department of Mycology and Plant Pathology (MPPL) and maintained in test tubes as slants and stored at 4℃. Seven different species of Aspergillus with their accession numbers, A. japonicus (503), A. niger (706), A. parasiticus (174), A. nidulans (722), A. flavus (647), A. oryzae (01) and A. fumigatus (651) were grown at 24-25℃.
2.2. Preparation of Culture
- Slants prepared in test tubes containing 5 ml of autoclaved medium (1 g yeast, 1 g peptone, 1 g starch and 1 g agar in 100 ml water) were inoculated with their respective cultures with the help of loop and incubated at 37℃ for 3-4 days. When growth appeared, they were stored at -20℃. A single colony from the starter culture was inoculated in 30 ml of autoclaved broth (0.01 g Fe SO4, 0.50 g KCl, 0.50 g Mg SO4, 1 g K2HPO4, 3 g NaNO3 and 20 g sucrose in one liter of water) in a 100 ml conical flask. Flask was covered with cotton plug and aluminum foil and placed in incubator for 4 days at 27℃. Growth appeared after one week.
2.3. RAPD PCR Amplifications
- Total DNA of fungal samples were extracted manually by CTAB method[15]. Optimized primer GL DecamerB-09: 5` TGGGGGACTC 3` and Primer GL DecamerB-10: 5` CTGCTGGGAC 3` were used for PCR of isolated DNA. 50 µl reaction mixture, containing 10X PCR buffer (5 µl), 5 mM dNTPs mix (2 µl), 5 U/µl Taq DNA polymerase (0.6 µl), template DNA (5 µl), 25 mM MgCl2 (4 µl), 50 pM Primer (1 µl) and PCR water (32.4 µl) each of it in a Bio-rad Thermal Cycler for 40 cycles, each for 1 min at 94℃, 1 min at 35℃ and 2 min at 72℃ and the final extension for 5 min at 72℃.
2.4. Statistical Analysis
- In order to access overall distribution of genetic diversity data was analyzed using Minitab software.
2.5. Biochemical Characterization
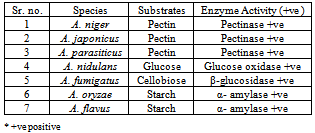
- For biochemical characterization Czapek Dox media (20 g agar, 20 g sucrose, 0.01 g FeSO4, 0.50 g KCl, 0.50 g Mg SO4, 01 g K2HPO4 and 3 g NaNO3) supplementd with 2% pectin for A. niger, A. japonicus and A. parasiticus, glucose for A. nidulans, starch for A. flavus and A. oryzae are added with starch to observe activity of α-amylase specifically and cellobiose for A. fumigatus respective substrate was inoculated with 5.106 spores/ml and incubated at 30℃ for 24 h. Plates were stained with Ruthenium red (0.05%) solution for 1 hour or with Congo red dye. Destaining was done by 1 M NaCl and tap water. Plates were observed for their biochemical and enzymatic reaction as by the formation of coloured zone in each case.
3. Results and Discussion
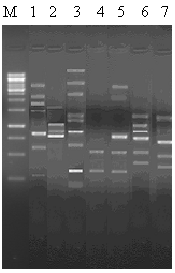
- Figures 1 and 2 shows polymorphism among seven Aspergillus species, which means a particular primer, gives different bands to evaluate species with minor genetic differences. A total of 46 bands with primer GL-Decamer B-09 (Figure. 1)
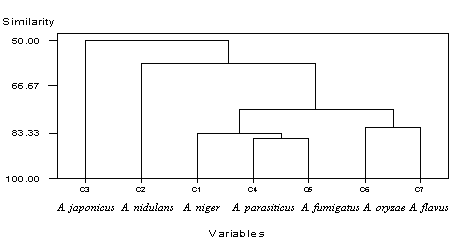 | Figure 3. Dandrogram showing genetic diversity with primer GL- Decamer B-09. C1, A. niger, C2, A. nidulans, C3, A. japonicus, C4, A. parasiticus, C5, A. fumigatus, C6, A. oryzae, C7, A. flavus |
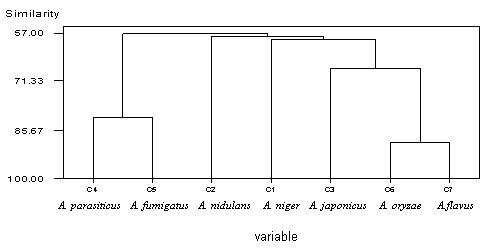 | Figure 4. Dandrogram showing genetic diversity with primer GL-- Decamer B-10. C1, A. niger, C2, A. nidulans, C3, A. japonicus, C4, A. parasiticus, C5, A. fumigatus, C6, A. oryzae, C7, A. flavus |
|
Acknowledgments
- We are thankful to Dr. Rukhsana Bajwa for providing us the fungal strains obtained from First Fungal Culture Bank of University of the Punjab Lahore at Institute of Mycology and Plant Pathology (IMPPL).
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-Text HTML
Full-Text HTML