-
Paper Information
- Previous Paper
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
Journal of Microbiology Research
2012; 2(2): 12-18
doi: 10.5923/j.microbiology.20120202.03
Antibiogram Status of Bacterial Isolates from Air Around Dumpsite of Ekiti State Destitute Centre at Ilokun, Ado-Ekiti, Nigeria
Odeyemi A. T
Department of Microbiology, Ekiti State University, Ado-Ekiti, Ekiti State, Nigeria
Correspondence to: Odeyemi A. T , Department of Microbiology, Ekiti State University, Ado-Ekiti, Ekiti State, Nigeria.
| Email: |  |
Copyright © 2012 Scientific & Academic Publishing. All Rights Reserved.
An investigation of the bacteriological quality of air around a municipal refuse dump in Ado-Ekiti, Nigeria was conducted to assess the bacteriological contamination of air and also to assess the ambient air quality of the closest neighbourhood to the dump site. The microbial concentration of air around the dump site was measured using the “passive method” that involved exposing sterile Petri dishes containing nutrient agar to the air for about five minutes. The exposures were carried out on separate days at some locations close to the dump site viz; the dumpsite, about 50m away from the dumpsite, and at the nearest neighbourhood which is about 100m away from the dumpsite. The microbes at the neighbourhood with mean value of 137.4 CFU/p was in most cases higher than the microbes at the dumpsite with mean value of 124.4 CFU/p. The bacterial distribution in the air revealed, 37% of Escherichia coli, 19% of Klebsiella spp, 13% of Pseudomonas spp, 15% of Serratia spp, 8% of Staphylococcus spp, 7% of Enterococcus spp and only 1% of Salmonella sp. The pattern of resistance of the bacterial isolates were Ceftazidime (87.7%), Cefuroxime (82.2%), Gentamicin (89.0%), Cefotaxime (84.9%), Ofloxacin (90.4%), Amoxicillin (95.9%), Augmetin (93.2%), Nitrofurantoin (93.2%) for the Gram negative bacteria, while, the percentage of resistant Gram positive isolates is as follows: Tetracycline (84.6%), Sulfamethoxazone (61.5%), Erythromycin (84.6%), Fusidic acid (69.2%), Gentamicin (61.5%), Clindamycin (30.8%), Penicillin (100%) and Trimethropin (92.3%). This research work revealed the relevance of an Environmental Microbiology Department in any Governmental Waste Management System and the potential hazard of the open dump system of waste disposal around residential area.
Keywords: Dumpsite, microbial, air contamination, waste management, resistance pattern
Article Outline
1. Introduction
- In Nigeria as well as in most developing countries, the urban landscapes are littered with garbage, plastics, bottles, disposable cups, discarded tires and even human and livestock faeces. These wastes are aesthetically unpleasant, constitute eyesores, produce unpleasant odour especially when their organic compositions are acted upon by putrefying bacteria. These refuse dumps thus constitute a habitat for vector and other nuisance organisms capable of transmitting or causing diseases such as typhoid, infantile diarrhoea and cholera in humans and animals (Siboe et al., 2006).Refuse dumps refer to areas or land sites where material wastes from several sources and processes are deposited. Refuse dumps include both municipal solid wastes and in dustrial wastes including liquid effluents containing heavy metals (Olanrewaju, 2002). Refuse dumps provide a rich source of microorganisms most of which are pathogenic (Odeyemi et al., 2011). This is usually as a result of the attraction of rodents and vector insects for which the dump serves as shelter and food source (Donderski et al., 2000). Although it is known that vector insects and rodents can transmit various pathogenic agents of diseases such as amoebic and bacillary dysentery, typhoid fever, salmonellosis, cholera, plague and so on. A good percentage of these infections are caused by bacteria which are suspended in air around these refuse dumps which may later settle and cause contamination. Activities involving the disposal of solid wastes even if properly controlled with proper precautionary measures adopted may have adverse impact on the environment especially air since most of the dumps are open.Microorganisms present in the refuse use the refuse as a food source. Under the anaerobic conditions typical in most dumps, these microorganisms convert the organic material in the refuse to methane and carbon dioxide. As the gas rises through the dump and escapes into the atmosphere, it sometimes picks up other compounds. The presence of large amounts of methane in this uncontrolled environment may result in explosions and fires. Additionally, this untreated gas may contain other compounds that pose a substantial health risk to nearby communities (Kerry et al., 2011).Many microbes can remain viable even after extended periods of time aloft despite the challenges associated with surviving in the atmosphere, including extended UV exposure, low moisture levels and extremely oligotrophic conditions (Jones and Harrison, 2004). Atmosheric transport is a key mode of microbial dispersal (Stetzenbach et al., 2004) and the transmission of airborne plant and animal pathogens can have significant impacts on ecosystems, human health and agricultural productivity.This study aimed at isolating bacteria present in air around a specific municipal dump in Ado-Ekiti metropolis, identifying the isolated organisms, and determining their level of resistance to antibiotics.
2. Materials and Methods
- Description of the study area and sampling locationThe study area is the Ekiti State Government Destitute Centre at Ilokun, Ado-Ekiti, used as dumping site by the Ekiti State Waste Management Board [EKWMB]. Most of the wastes disposed are mainly domestic and household wastes such as food residues from kitchen, hair and dead skin cells from bath/shower water, and human excreta (urine and faeces).Collection of samples and bacteriological analysisThe isolates were collected from different locations in and around the dump. These locations included the dump site A, B (150m to the dumpsite; the closest point to neighborhoods), C (50m away from the dumpsite), and a nearby stream D (100m away from the dumpsite). Twenty millilitre (20ml) of nutrient agar medium was poured into sterile Petri-dishes and allowed to solidify. The plates were then sealed and labeled appropriately and taken to the dump site where each was exposed for about five minutes at the selected locations. The plates were afterwards covered and taken into the laboratory for incubation at 37oC for 48hrs. This procedure was repeated for five consecutive days.The pure bacterial strains were identified on the basis of their morphological and biochemical tests. The pure cultures of the bacterial isolates were subjected to various morphological and biochemical characterization tests such as color, shape, elevation, consistency , margin , catalase test, MRVP (Methyl Red-Voges Proskauer test), fermentation of sugars, kovacs citrate, indole, hydrolysis of starch, and sensitivity tests (Olutiola, 1991). In order to determine the identity of bacteria isolates, results were compared with standard references of Bergey’s Manual of Determinative Bacteriology 2nd edition (Buchanan and Gibbons, 1974).Antibiotic susceptibility testThe antibiotics susceptibility of the isolates was determined by the disk diffusion method on Mueller-Hilton agar according to CLSI (2005). Bacterial isolates were tested against seven ABTEK disc antibiotics which comprised Cefotaxime (CAZ 30µg), Cefuroxime (CRX 30µg), Gentamycin (GEN 10µG), Ofloxacin (OFL 5µg), Augmentin (AUG 30µg). Gram negative disc contains additional constituent such as Nitrofurantoin (NIT 300µg), Ceftazidme (CTX 30µg) and Amoxicillin (AMX 30 µg). Gram positive disc contains additional constituent such as Lincomycin (LIN 2µg), Oxacilin (OXA 10µg) and Cloxacilin (COX 5µg). The inoculum was standardized by adjusting its density to equal the turbidity of a barium sulphate (BaSO4) (0.5 McFarland turbidity standard), and incubated at 35oC for 18 hours. The diameter of the zone of clearance (including the diameter of the disk) was measured to the nearest whole millimeter and interpreted on the basis of CLSI guideline (CLSI, 2005).Statistical analysisThe results are expressed as Mean ±SD. Difference in means were also determined by Duncan’s multiple range test (P<0.05).
3. Results and Discussion
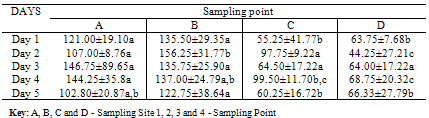
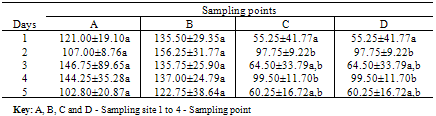
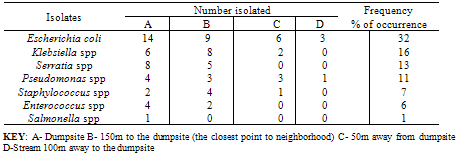
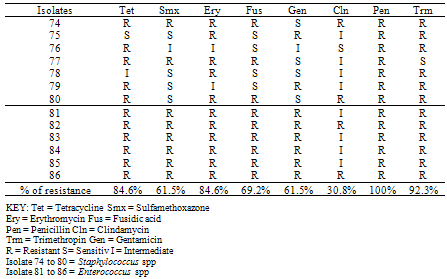
- The mean values for the total bacterial counts ranged as follows: A (124.4 CFU/p), B (137.5 CFU/p), C (75.5 CFU/p) and D (61.4 CFU/p) with the neighborhood having the highest number of counts which are closely followed by the dumpsite, and the stream having the least number of counts. The Duncan Multiple Range Test, performed on the total bacterial counts obtained at the different sampling locations on five separate days showed the mean value for each count as well as the standard deviation, the variation between the different days of sample collection and locations at 0.05 levels (Table 1a & b).On day one, there is no significant variation between the bacterial counts at locations C and D, and between locations A and B respectively, but the counts at location C and D are significantly different from those counts from locations A and B. In day 2, bacterial counts at locations A and C are significantly similar, but dissimilar to the bacterial counts at locations B and D. In day 3, the counts obtained in all the sites are significantly similar. On the 4th day, the microbial counts at the locations A and B are similar, B and C are similar, C and D are also similar but the count at location A differs significantly from location C, B from D, and A from D. The result on the 5th day shows that the bacterial counts at location A is similar to that of locations B, C and D, C and D are similar, but counts at location B varies from locations C and D. All counts are significantly similar at sites A and B throughout the collection period. Also, the bacterial counts obtained on the 1st day are similar to the counts obtained on the 3rd and 5th days, while the counts obtained on 2nd and 4th days are significantly similar to the counts obtained on the 3rd and 5th days (Table 1b).According to the bacterial distribution in the air, 37% were Escherichia coli, 19% were Klebsiella spp, 13% were Pseudomonas spp, 15% were Serratia spp, 8% were Staphylococcus spp, 7% were Enterococcus spp and only 1% was Salmonella sp. Escherichia coli had the highest frequency, this was followed by Klebsiella spp., Serratia spp was next in the frequency of occurrence, followed by Pseudomonas spp, Staphylococcus spp, Enterococcus spp and Salmonella spp having the least frequency of occurrence (Table 2).The pattern of resistance of the bacterial isolates were Ceftazidime (87.7%), Cefuroxime (82.2%), Gentamicin (89.0%), Cefotaxime (84.9%), Ofloxacin (90.4%), Amoxicillin (95.9%), Augmetin (93.2%), Nitrofurantoin (93.2%) for the Gram negative bacteria (Table 3). While, the percentage of resistant Gram positive isolates is as follows: Tetracycline (84.6%), Sulfamethoxazone (61.5%), Erythromycin (84.6%), Fusidic acid (69.2%), Gentamicin (61.5%), Clindamycin (30.8%), Penicillin (100%) and Trimethropin (92.3%) (Table 4).
|
|
|
|
4. Discussion
- The results obtained in this study correlates with that of Noah et al., (2008). The result shows that the mean total bacterial counts obtained from the dumpsite and the residential area close to the dumpsite were relatively higher than those obtained at the stream and 50m away from the site respectively, with the bacterial count obtained from the residential area having the highest value.The high value of bacteria which are majorly pathogens in the residential area can be attributed to its nearness to the dumpsite as well as the various and diverse human activities in the area which include rearing animals, raising dust through walking and various farming practices, fermentation activities, uncontrolled disposal of solid wastes and sewage, unethical faeces disposal and so on. The dumpsite as well on its own offers a rich source of organic matter and factors that favour the abundance of these organisms which are occasionally raised from the dump into the air by wind action.The decrease in microbial load as we move away from the dumpsite, precisely, 50m away from the dumpsite can be attributed to the antimicrobial property of the ultraviolet light and solar radiation of the sunlight and decrease in the quantity of organic matter present for microbial use. Also, the stream, having the least mean value of bacterial count can also be attributed to the solar radiation of the sunlight and also, the natural buffer and filter systems in water, also since the stream is slow flowing and has no tidal action; therefore, the displacement of organisms from it is minimal.From the statistical analysis, it can be deduced that only a little variation exists between the bacterial counts obtained between different days at the same location and at the different locations on the same day of sample collection. This is because the study was conducted within a limited range of time that does not permit enough room for seasonal variation.The bacterial species isolated were identified to be among those commonly encountered in leachate and runoff from the waste pile (Obire et al., 2002). With the exception that the other few found in leachate but not encountered in this study may be due to their survival and settling time. The high number of bacteria from the family Enterobacteriaceae and considerable number of coliform bacteria is an indication that the environment is hazardous and constitutes serious health risk and threat to both the waste workers and residents of the nearby community. According to Scarpino et al., (2010), people in close proximity to the dumpsite complained of serious odours emanating from the site, personal discomfort due to the odours, loss of sleep, possible allergic manifestations and respiratory difficulties. Out of the genera of organisms isolated, Escherichia coli has the highest frequency of occurrence, this agrees with the reports of Lewis et al., (2002) which stated that Escherichia coli is able to withstand competition from the other indigenous microorganisms with higher growth rates.From the results obtained in this study, antibiotics resistant bacteria were widespread as nearly all the isolated organisms were resistant to most of the antibiotics for which they were tested against. This may be due to either the intrinsic resistance of many microorganisms to antibiotics or acquired resistance of the organisms enabled by the transfer of resistance of drug resistance plasmids among members of the isolates. A high level of resistance has been found with members of the family Enterobacteriaceae which are believed to have increased the incidence of pathogenic strains of bacteria with acquired antibiotics resistance. The origin of this resistance can be traced to the faecal constituent of the wastes or dump produced by people or animals that have been treated indiscriminately with various antibiotics and also to antibiotics production naturally by soil microorganisms (Ajayi et al., 2003). Since antibiotics in animal feeds promote animal growth, improved efficiency of feed conversion to body weight, and may also affect disease prophylaxis among the confined microbes in dosed animals and their subsequent impact on human health, it has increased its indiscriminate use (Scarpino et al., 2010).
5. Conclusions
- From this study, it can be concluded that the open dump system of waste disposal is indeed a potential environmental quality problem which takes the form of unsightliness, land and water pollution, it reduces the quality of air by the emission of foul odours and different gases derived from the anaerobic decomposition as well as occasional burning. It also serves as a potential source of air pollution and contamination as it promotes the dispersion of bacterial pathogens either as free entities or attached to particles into the air. These pathogens when suspended in air are of less importance but become a source of immediate concern when they settle on surfaces as they cause varying kinds of infectious diseases, respiratory symptoms and lung function impairment which can range from acute mild conditions that hardly affect daily life to severe chronic respiratory diseases, cancer, and so on, that require specialist’s care.In order to enhance the quality of air and protect the lives of people, the following recommendations need to be considered and implemented.i. A land fill waste disposal system should replace the open system of waste disposal as this will ensure the effective control and prevention of microorganisms from escaping into the air.ii. In case of limited land availability, the wastes can be incinerated under high heat in a controlled environment.iii. Encouragement of waste management practices of waste reduction, waste re-use and recycling. Government should also encourage small and medium scale industries that can convert these wastes into useful products by creating enabling environment.iv. Legislative laws and regulations on land use and effective waste disposal and management should be made to control the location of dumps which should be far removed from the community.v. Hazardous wastes from industries and hospitals should be treated or detoxified before disposal.vi. Government should control the settlement patterns of individuals and communities and ensure that residential are far removed from dumpsites.vii. Public health organisations and other relevant bodies should embark on public awareness and enlightenment campaigns to enlighten individuals on the hazards of indiscriminate waste disposal and the open dump system of waste disposal.
References
| [1] | Ajayi, A.O. and Akonai, K.A. (2003). Antibiotics sensitivity profile of microorganisms in Lagos lagoon, Nigeria. African journal of Biotechnology 6: 79-84. |
| [2] | Brodie, E., DeSantis, T.Z., Parker, J., Zubietta, I., Piceno, Y. and Anderson, G. (2007). Urban aerosols harbour diverse and dynamic bacterial populations. Proc. National Academy of Science. USA. 104: 299-304. |
| [3] | Buchanan, R.E and Gibbons, N.E. (1974). Bergey’s Manual of Determinative Bacteriology 8th edition. The Williams and Wilkins company, Baltimore. |
| [4] | CLSI, Performance standards for antimicrobial susceptibility testing; fifteenth informational supplement, Clinical and Laboratory Standard Institute Wayne, Pa. M100-S15, 2005, vol.25, no.1. |
| [5] | Jones, A. And Harrison, R. (2004). The effects of meterological factors on atmospheric bioaerosol concentrations: a review. Sci. Total Environment. 326: 151-180. |
| [6] | Lewis, D.L. and Gattie, D.K. (2002). Pathogen risks from applying sewage sludge to land. Environmental science Technology Journal. 36: 286A-293A. |
| [7] | Maleicka-Adamowicz, M., Kaczanowska, J. And Donderski, W. (2007). The impact of a landfill site in Zolwin- Wypaleniska on the microbiological quality of the air. Polish journal of Environmental studies. Vol. 16, No. 1.30:101-107. |
| [8] | Matthias-Maser, S., Obolkin, V., Khodzer, T. and Jaenicke, R. (2000). Seasonal variation of primary biological aerosol particles in the remote continental region of lake Baikal/Siberia. Atmospheric Environment. 34: 3805-3811. |
| [9] | Monica, C. (2000). District laboratory practice in tropical countries. Journal of Microbiology. 2(7): 178-179. |
| [10] | Noah, F., Zongzhi, L., Mari, R., Rob, K., Matthew, H. and Mark, T.H. (2008). Short-term temporal variability in airborne bacterial and fungal populations. Applied and Environmental Microbiology. Volume 74, No.1. Page 200-207. |
| [11] | Obire, O., and Aguda, M. (2002). Bacterial community of leachate from a wastedump and adjacent stream. Journal of applied sciences and environmental management, volume 6, number 2, pp. 71-75. |
| [12] | Odeyemi, A.T., Faweya, E.B., Agunbiade, O.R and Ayeni, S.K. (2011). Bacteriological, mineral and radioactive contents of leachate samples from dumpsite of Ekiti State Government Destitute Centre in Ado-Ekiti. Archives of Applied Science Research, 3 (4):92-108. |
| [13] | Olanrewaju, A.O. (2002). Dangers of indiscriminate refuse dumps in metropolis countries. Environment opinions. 35: 567-571. |
| [14] | Olutiola, P.O., Famurewa, O and H.E. Sontag, H.E. (1991). An introduction to General Microbiology, a practical Approach Heideberger Verlagsanstalt and Druckerei GmbH Heldelberg Gmbh, Germany. |
| [15] | Onyido, A.E., Okolo, P.O., Obiukwu, M.O., and Amadi, E.S. (2009). A survey of vectors of public health diseases in undisposed refusedumps in Akwa town, Anambra state, south eastern Nigeria. Research Journal of Parasitology, 4:22-27. |
| [16] | Pillai, S.D., and Ricke, S.C. (2002). Canadian Journal of Microbiology. 48(8):681-696 (ISSN: 0008-4166). |
| [17] | Sattler, B., Puxbaum, H., and Psenner, R. (2001). Bacterial growth in supercooled cloud droplets. Geophys. Res. Lett. 28:239-242. |
| [18] | Scarpino, P.V., and Quinn, H. (2010). Department of civil Engineering, the University of Cincinnati, Cincinnati, Oklahoma. 45221-0071. |
| [19] | Stetzenbach, L., Buttner, M. And Cruz, P. (2004). Detection and enumeration of airborne biocontaminants. Curr. Opin. Biotechnology. 15:170-174. |
| [20] | Wathes, C.M. and Cox, C.B. (1995). Bioaerosols handbook. Chelsea, Mich: Lewis Publishers. ISBN 0-87371-615-9. |
| [21] | Womiloju, T.O., Miller, J.D., Mayer, P.M. and Brook, J.R. (2003). Methods to determine the biological composition of particulatematter collected from outdoor air. Atmospheric Environment. 37:4335 - 4344. |
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-Text HTML
Full-Text HTML