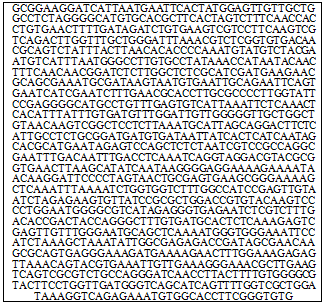-
Paper Information
- Next Paper
- Previous Paper
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
Journal of Microbiology Research
p-ISSN: 2166-5885 e-ISSN: 2166-5931
2012; 2(1): 11-17
doi: 10.5923/j.microbiology.20120201.03
Improvement of Laccase Production in Pluerotus pulmonarius-LAU 09 by Mutation
Adebayo Elijah. Adegoke 1, Oloke Julius. Kola 1, Achana Yadav 2, Bora Tarun. Chandral 2
1Department of Pure and Applied Biology, Ladoke Akintola University of Technology, P.M.B. 4000, Ogbomoso, Nigeria
2North East Institute of Science and Technology, CSIR, Jorhat, Assam, 785006, India
Correspondence to: Adebayo Elijah. Adegoke , Department of Pure and Applied Biology, Ladoke Akintola University of Technology, P.M.B. 4000, Ogbomoso, Nigeria.
| Email: |  |
Copyright © 2012 Scientific & Academic Publishing. All Rights Reserved.
The strain improvement of P. pulmonarius LAU 09 for laccase production was carried out by UV- light exposure at 210nm (UV-sterilizer, Millipore xx63 70000) for 90minutes. The highest laccase activity of 2.5 Uml-1 was produced by LAU 90 (mutant) in comparison with obtained yield by wild type LAU 09 (1.75 Uml-1). The optimal physiological parameters for optimum laccase production by LAU 90 are 30OC of temperature, pH of 5.0 and incubation of 168hours. The mutant strain gave the highest total enzyme activity and enzyme specific activity of 71.7 x 10-4 mol/min and 89.63X 10-4 IU/mg of purified laccase enzyme respectively, with molecular weight of 97KDa. The activity of the purified enzyme was strongly inhibited by Sodium azide and EDTA at 1ml/0.1mM and 0.5ml/100mM respectively. The result obtained shown an improved performance of the mutant (LAU 90) strain of P. pulmonarius in laccase production over the wild (LAU 09) type.
Keywords: Strain, Improvement, Mutation, Laccase, Enzyme
Cite this paper: Adebayo Elijah. Adegoke , Oloke Julius. Kola , Achana Yadav , Bora Tarun. Chandral , "Improvement of Laccase Production in Pluerotus pulmonarius-LAU 09 by Mutation", Journal of Microbiology Research, Vol. 2 No. 1, 2012, pp. 11-17. doi: 10.5923/j.microbiology.20120201.03.
1. Introduction
- Laccases are multinuclear copper-containingglycoproteins that belong to the family of enzymes known as oxidases, more specifically “blue” oxidases[25], and polyphenol oxidases[8]. Laccases are one of six enzyme classes capable of reducing dioxygen to water, five of which belong to the multicopper oxidase family (the only enzyme not in this class being cytochrome-c oxidase, a heme/copper containing enzyme). Laccase is a polyphenol oxidase, indicative of the fact that laccases can oxidize a phenolic substrate that in turn can initiate a polymerization reaction[8]. Laccases from various sources vary greatly with respect to their degree of glycosylation, molecular weight and kinetic properties[25].Basidiomycetes fungi especially Pleurotus species are the most efficient lignin-degrading organisms that produce mainly laccases (EC 1.10.3.2), lignin peroxidase (EC 1.11. 10.14) and manganese peroxidase (EC1.11.1.13). These enzymes present a non-specific biocatalyst mechanism and have been used for bioremediation process due to their ability to degrade azo, heterocyclic, reactive and polymeric dyes[1, 6]. Prospection for fungi is the ability to secret high levels of lignin-degrading enzymes and novel enzyme variants, with desirable properties for biotechnological applications. On the other hand, alternative low cost substrates like agricultural residues for enzyme production using solid state fermentation (SSF) offer economic and environmental advantages. The main technological applications of laccases are in the textile, dye or printing industries, in processes related to decolouration of dyes[4], in the pulp and paper industries for the delignification of woody fibres, particularly during the bleaching process[3,13]. In most of these applications, laccases are used together with a redox mediator. At the end of 2005 three industrial processes were using laccases: dye bleaching, lignin bleaching and bleaching of cork for bottled wine[19].Laccases exhibit a broad natural substrate range, which is a major reason for the attractiveness of laccases to biotechnological applications[16]. Even more interesting however is the application of laccase activity to a broader substrate range through the secondary activity of the free (cation) radical formed by oxidation of its substrates. The substrates that result in this type of activity towards other compounds are termed mediators. The industrial applicability of laccase may therefore be extended by the use of a laccase-mediator system. In view of the importance of laccase mentioned above, the improve production of the enzyme is inevitable.Ultra violent radiation (UV- light) has been reported as one of the best physical method of strains improvement for better yield performance[10]. This method has been employed in improving enzyme production in Aspergillus niger[10], Rhizopus oryzea[22], mycelia cell and sporophore production in P. florida and P. sajor-caju[20]. However, no report is available on the improvement of laccase production by P. pulmonarius using UV- light radiation exposure. The present investigation was undertaken to improve the laccase production by P. pulmonarius - LAU 09 strain through exposure to UV-light radiation.
2. Material and Methods
- Mutation InductionThe actively growing culture (7 days old) of P. pulmonarius LAU 09 on PDA plates (90mm) were exposed to UV-light (210 nm, Millipore xx63 70000) for 90mins. The mutants were subcultured on the PDA with 5% of yeast extract agar (YEA), incubated at 25℃ for 7days.Enzyme AssaysThe strains, both LAU 09 (wild) and LAU 90 (mutant) were grown in PDB with 5% of yeast extract (YE). The experiments were carried out in 250ml Erlenmeyer flask with 50ml of substrate, inoculated with a plug (6mm), incubated at 25℃ for 5days at 150rpm. The culture were sieved (Whatman paper II), centrifuged and filtrate were used as crude enzymes. Laccase activity was determined via the oxidation of 2, 2-azino-bis (3-ethylbenzthiazoline)- 6- sulfonate (ABTS) (Sigma, America). The reaction mixture containing 0.1ml of 0.3mM ABTS in 100mM of Sodium acetate (pH 3.5) and 0.1ml of crude enzyme solution was incubated at 40℃ for 1min. The ABTS oxidation was monitored by the increased in absorbance at 420nm (є = 36000M-1 cm-1). One unit was defined as 1µmol of ABTS oxidized per minute and activity was expressed in Uper ML per min[15].Effect of temperature on enzymes productionThe optima temperature for laccase enzymes for LAU 09 and LAU 90 strains were evaluated by incubating the enzymes at different temperature ranges (25℃, 30℃, 35℃, 40℃, 45℃ and 50℃) with appropriate substrates and buffers as described above.Effect of pH on enzymes productionThe optima pH for laccase was determined by incubating the enzyme with their appropriate substrates at their optimum temperature using Mcllvaine’s method[17].Effect of incubation period on enzymes productionThe optimum incubation period for laccase production was determined by incubating the enzyme with their appropriate substrate at their optimum temperature for 9days[17].Acetone precipitationThe crude enzymes (20% w/v), 10,000g supernatant was subjected to 80% acetone precipitation at -4°C. The contents were then placed in ice for an hour and then centrifuged at 10,000g for 30 min at -4°C. The pellet was re-dissolved in 0.1 M glycine–NaOH, pH 10.0.Ion exchange column chromatographyAliquots (3 mL) of the acetone precipitated proteins were applied to a column of Sephadex A-50 ion exchange resin (30 X 0.8 cm) equilibrated with 0.1 M citrate buffer, pH 5. The protein sample volume was adjusted to 10% of the column bed volume. The ion exchangeChromatography of the proteins was carried out at room temperature (35℃). Following sample application, the column was washed with the equilibrating buffer, until the absorbance of the wash at A280 was <0.05. The adsorbed proteins were then eluted using a linear gradient of NaCl (0–1.0 M) in 0.1 M glycine–NaOH buffer, pH 10.0 at a flow rate of 0.9 mL/min. Fractions of 2mL were collected manually. Protein content of each fraction was evaluated at A280 employing a UV–vis double beam spectrophotometer (Spectra scan UV 2700: Thermo Scientific). Laccase activity in the fractions was determined using the enzyme assay as described above.Sephadex G-100 gel permeation column chromato- graphySephadex G-100 was allowed to swell in 0.1 M glycine–NaOH buffer, pH 10.0 for 72 h at room temperature and then packed on to a glass column (30 X 0.8 cm) with a flow rate of 0.9 mL/min. Active fractions of ion exchange chromatography were loaded onto the column. Fractions of 2mL were collected and the absorbance in each fraction was measured at 280 and 469 nm for the enzyme activity. The molecular weight was determined using 10% sodium dodecyl sulfate – polyacryl-amide gel electrophoresis (SDS- PAGE).Determination of protein concentrationThe concentration of protein was determined by Lowry’s Method[14].Laccase enzyme inhibitor StudiesThe effects of Sodium azide and EDTA on purified enzyme activity were tested using ABTS as substrate after pre-incubating the enzyme for 10mins at 25℃ with inhibitors before addition of substrate[21].SDS-PAGEMolecular weight of the purified enzyme was determined under the reducing condition of the SDS-PAGE according to Laemmli method[12].DNA extraction and ITS amplificationMycelia were grown on potato dextrose agar, harvested using a scalpel, transferred into Epperdorf tubes, small amount of autoclaved refined sand (Sigma) was added and ground to fine paste with pestle-like stick (High Media), 400µl of DNA Extraction buffer pH 8 (1M Tris-Cl pH 8.0; 1M NaCl; 200mM EDTA pH 8.0; 10%SDS; 0.1%β- Mercaptoethanol) was added and centrifuged at 4℃ (12000g) for 10mins. To the collected supernatant 300µl Phenol and 300 µl Chloroform: Isoamylalcohol (24:1) were added and mixed gently. This was centrifuged (12000g, 4℃ for 10mins), and aqueous phase was collected and 500µl chilled Isopropanol was added and incubated at -20℃ overnight. After the incubation, it was centrifuged (12000g, 4℃ for 10mins), and the pellet was washed with chilled 70% ethanol centrifuged for 5mins. The dried pellet was resuspended in 50 µl of Tris EDTA (10Mm Tris and 1mM EDTA, pH 8.0) buffer.Amplification of the ITS region of the rRNA gene was carried out with a modified method of Gerdes and Bruns[7], using primers ITS1-F and ITS4-B. The final concentration of 25µl PCR reaction volume were; 200 µM each of dATP, dCTP, dGTP and dTTP, 2.5mM MgCl2, 10X Taq.DNA Polymerase and 20Pico mole of each of the two primers (Banglore Genei). The PCR profiles were initial denaturation step of 94℃ for 85s followed by 25 amplification cycles of denaturation, annealing and extension. The temperature and times for these steps were 95℃ for 35s, 55℃ and 55s and 72℃ for 2mins with further incubation at 72℃ for 10mins. The amplified PCR products were resolved on a 1.2% agarose gel, and stained with Ethidium bromide. A 1kb ladder DNA marker (GeneRulerTM ) was used as a size standard.Sequencing and Phylogenetic AnalysisThe PCR products were purified using Exonuclease I and Shrimp Alkaline phosphatasa in buffer (EXOSAP Kits). Both strands of the amplified region were sequenced using fluorescent dye terminator chemistry and were run on ABI 3130 (4 capillary) or 3730XI (96 capillary) Automated Sequencer (Perkin Elmer Applied Biosystems, Foster City, CA), following the manufacturer’s protocols. Sequencing primers were ITS1-F, 5.8S, 5.8SR and ITS4-B. Oligonucleotide sequences for primers 5.8S and 5.8SR were given in Vilgaly and Hester[23]. Sequence contigs were assembled and edited using Sequencer 3.0 software (Gene codes Corporation, Ann Arbor, MI).Phylogenetic trees were constructed by using all cloned sequences together with all non redundant large subunit (nLSU) sequences of named Pleurotus species obtained from GenBank. The multiple alignments of all the sequences were performed using CLUSTAL W(http://www.ebi.ac.uk/Tools/msa/clustalw2/), followed by manual adjustments. The phylogenetic analyses was carried out using sequence data of ITS 5.8s and 28s ribosomal RNA gene from LAU 09 (wild) of P. pulmonarius and corresponding GenBank data of related species. The identical sequences were merged into one input sequence when running the computer programs to generate the phylogenetic trees constructed by UPGMA, Neighbor-joining (NJ) and parsimony methods. The boostrap test for estimating the reliability of phylogenetic tree topology was performed using 100 replications by the SEQBOOT program[5]. The consensus tree was obtained by running the consense program[5].
3. Results and Discussion
- The laccase enzyme activity of the wild and mutant strains of P. pulmonarius was shown (Figure 1), with highest yield (2.5 Uml-1) obtained by LAU 90 (mutant) comparing with (1.75 Uml-1) obtained by LAU 90 (wild). The results obtained by the LAU 90 strain have shown an improved enzymes production over wild type (LAU 09).Th The highest production of laccase, enzyme could be as result of efficient lignin-degrading ability of P. pulmonarius. Basidiomycetes fungi are the most efficient lignin-degrading organisms that produce mainly laccases, manganese peroxidase, and lignin peroxidase[9]. These enzymes present a non-specific biocatalyst mechanism and have been used for bioremediation process due to their ability to degrade azo, heterocyclic, reactive and polymeric dyes[1,6]. The increased yield in laccase production by LAU 90 is an evidence of true strain improvement. This correlate with work of Kang et al[10], who reported an improving enzyme production in Aspergillus niger through UV-light mutation, Rhizopus oryzea[22], mycelia cell and sporophore production in P. florida and P. sajor-caju[20].
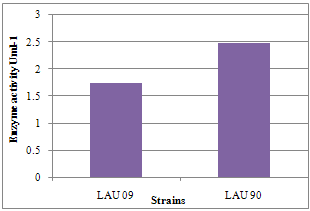 | Figure 1. Laccase activity of wild and mutant strains of Pleurotus pulmonarius |
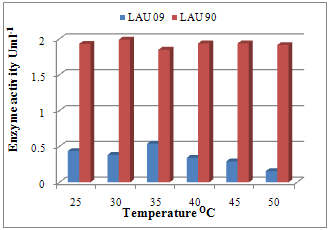 | Figure 2. Effect of temperature on laccase activity of wild and mutant strains of P. pulmonarius |
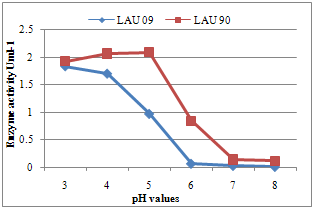 | Figure 3. Effect of pH on laccase activity of wild and mutant strains of P. pulmonarius |
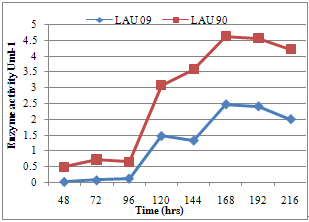 | Figure 4. Effect of incubation period on laccase activity of wild and mutant strains of P. pulmonarius |
|
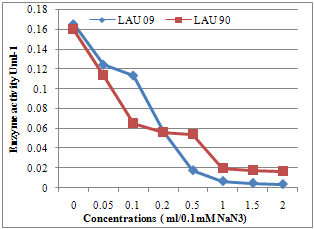 | Figure 5. The lacccase enzyme inhibition test by Sodium azide |
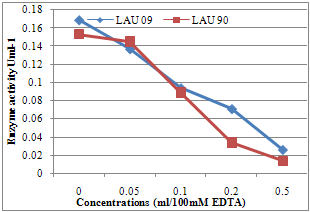 | Figure 6. The lacccase enzyme inhibition test by EDTA |
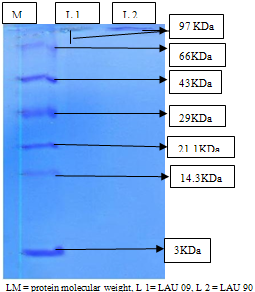 | Figure 7. Protein (laccase) Molecular Weight as visible on 10 % SDS-PAGE after staining with Coomassie Brilliant Blue |
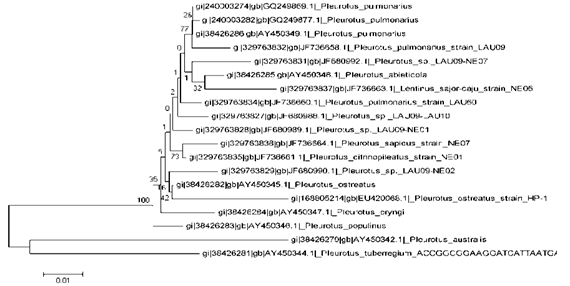 | Figure 8. The Phylogenetic trees of LAU 09 (wild) constructed by UPGMA, with boostrap values from 100 replicates showing maximum similarity with LAU 09 (wild) and the homogeneous strains of Pleurotus species |
|
|
ACKNOWLEDGMENTS
- The Director of NEIST, India is gratefully acknowledged for granting facilities available to carry out this research, so also TWAS, Italy; and CSIR for the award of Postgraduate Fellowship given to me (Adebayo, E.A) and utilized at NEIST, CSIR, Jorhat, India, and also to the authority of LAUTECH, Ogbomoso, Nigeria for granting the study leave to utilize the award.
References
| [1] | Baldrian, P., and Snajdr, J., 2006, Production of ligninolytic enzymes by litter-decomposing fungi and their ability to decolorize synthetic dyes. Enzyme Microbiol. Technol 39: 1023-1029 |
| [2] | Bao, D., Ishihara, H., Mori, N., Kitamoto, Y., 2004, Phylogenetic analysis of oyster mushrooms (pleurotus spp.) based on restriction fragment length polymorphisms of the 5’ portion of 26s rDNA. J wood sci, 50: 169-176 |
| [3] | Bajpai, P., 1999, Application of enzymes in the pulp and paper industry. Biotechnol Prog 15:147–57 |
| [4] | Claus, H., Faber, G., König, H., 2002, Redox-mediated decolorization of synthetic dyes by fungal laccasse. Appl Microbiol Biotechnol 59:672–678 |
| [5] | Felsenstein, J., 1989, PHYLIP –Phylogeny Inference package (version 3.2). Cladistic 5:164-166 |
| [6] | Forgacs, E., Cserháti, T., Oros, G., 2004, Removal of synthetic dyes from wastewaters: a review. Environ. Intern, 30, 953-971 |
| [7] | Gerdes, M., and Bruns, T. D., 1993, ITS primers with enhanced specificity for basidiomycetes- application to the identification of mycorrhizea and rusts. Molecular Ecology 2: 113-118 |
| [8] | Gianfreda, L., Xu, F., Bollag, J. M., 1999, Laccases: A useful group of oxidoreductive enzymes. Biorem. J. 3:1-25 |
| [9] | Gomes, E., Aguiar, A. P., Carvlho, C. C., Bonta, R. M., Desilva, R., Boscolo, M., 2009, Ligninases production by Basidiomycetes strains on lignocellulosic Agricultural residues and their application in the decolorization of synthetic dyes. Brazillian journal of Microbiology 40:31-39 |
| [10] | Kang, S .W., Ko, E. H., Lee, J. S., Kim, S.W., 1999, Overproduction of beta-glucosidase by Aspergillus niger mutant from lignocellulosic biomass. Biotechnol. Lett 21: 647- 650 |
| [11] | Kumar, G.N., and Srikumar, K., 2010, Thermophilic Laccase from xerophytes species Opuntia vulgaris. Research article (Wileyonlinelibrary.com) DOI 10.1002/bmc.1506 |
| [12] | Laemmli, U. K., 1970, Cleavage of structural proteins during assembly of head of bacteriophage T4. Nature 227: 680-685 |
| [13] | Leonowicz, A., Cho, N.S, Luterek, J., Wilkolazka, A., Wojtas-Wasilewska, M., Hofrichter, M., 2001 |
| [14] | Fungal laccase: properties and activity on lignin. J Basic Microbiol, 41:185–227 |
| [15] | Lowry, O.H., Rosebrough, N.J., Farr, A.L., Randall, R.J., 1951, Protein measurement with the folin phenol reagent. J. Bio. Chem 193.265-275 |
| [16] | Machado, K. M. G., and Matheus, D. R., 2006, Biodegradation of remazol brilhant blue R by ligninolytic enzymatic complex produced by Pleurotus ostreatus. Braz. J. Microbiol, 37: 468-473 |
| [17] | NYANHONGO, G. S., GOMES, J., GÜBITZ, G., ZVAUYA, R., READ, J. S., STEINER, W., 2002, Production of laccase by a newly isolated strain of Trametes modesta. Biores. Technol 84, 259–263 |
| [18] | Okamoto, K., Yanagi, S. O., Sakai, T., 2000, Purification and characterization of extracellular laccase from Pleurotus ostreatus. Mycoscience, 41: 7-13 |
| [19] | Pang, Q., Zhang, S., Shi, X., Shu, F., Wu, D., 2005, Purification and characterization of phenoloxidase from amphioxus Branchiostoma belcheri. Tsingtauense, 19: 139-148 |
| [20] | Riva, S., 2006, Laccases: blue enzymes for green chemistry. Trends Biotechnol 24:219–226 |
| [21] | Sandhya, R., Meera, P., Tewari, R.P., Krishna, V., 2006, Development of sporeless/low sporing strains of Pleurotus through mutation. World Journal of Microbiology & Biotechnology 22: 1021- 1025 |
| [22] | Soden, D. M., Callaghan, J. O., Dobson, A. D. W., 2002, Molecular cloning of a laccase isozyme gene from Pleurotus sajor-caju and expression in the heterologous pichia pastoris host. Microbiology 148: 4003-4014 |
| [23] | Suntornsuk, W., Hang, Y. D., 2008, Strain improvement of Rhizopus oryzae for production of (+)- lactic acid and glucoamylase. Lett. Appl. Micobiol 19: 249-252 |
| [24] | Vilgalys, R., and Hester, M., 1990, Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol 172, 4238-4246 |
| [25] | Vilgalys, R., Moncalvo, J. M., Liou, S.R.,Volovcek, M., Recent advances in molecular systematics of the genus Pleurotus. In Mushroom Biology and Mushroom Products, pp. 91–102. Edited by D. J. Royse. PennState: PennState University, 1996 |
| [26] | Yaropolov, A. I., Skorobogat’ko, O. V., Vartanov, S. S., Varfolomeyev, S. D., 1994, Laccase: Properties, Catalytic Mechanism, and Applicability. Appl. Biochem. Biotechnol 49:257-280 |
| [27] | Zervakis, G., Philippoussis, A., Ioannidou, S., Diamantopoulou, P., 2001, Mycelium growth kinetics and optimal temperature conditions for the cultivation of edible Mushroom species on Lignocellulosic substrates. Folia Microbiol, 46: 3: 231- 234 |
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-Text HTML
Full-Text HTML