-
Paper Information
- Next Paper
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
Basic Sciences of Medicine
p-ISSN: 2167-7344 e-ISSN: 2167-7352
2012; 1(1): 1-5
doi: 10.5923/j.medicine.20120101.01
PCR Based Detection of Parental Origin of Extra Chromosome 21 in Children with Down Syndrome in Pakistan
Tooba Altaf , Saba Irshad
Institute of Biochemistry and Biotechnology, University of the Punjab, Quaid-e-Azam campus, Lahore, 54590, Pakistan
Correspondence to: Saba Irshad , Institute of Biochemistry and Biotechnology, University of the Punjab, Quaid-e-Azam campus, Lahore, 54590, Pakistan.
| Email: |  |
Copyright © 2012 Scientific & Academic Publishing. All Rights Reserved.
Down syndrome, the most frequent form of mental retardation caused by a microscopically demonstrable chromosomal aberration, is characterized by well-defined and distinctive phenotypic features. It is caused by triplicate state (trisomy) of all or a critical portion of chromosome 21. The risk of having a child with trisomy 21 increases with maternal age. Out of the total thirty patients 53.3% were males and 46.6% were females. Down syndrome is slightly common in male children than in females. The parental origin of the extra chromosome has been studied in thirty families each with a clinically suspected trisomy 21 proband. Molecular analysis was carried out by PCR based method, using polymorphic microsatellite markers D21S11 and D21S2055 situated on the long arm of the chromosome 21 at 21q21 and 21q22 respectively. The amplified products were subjected to polyacrylamide gel electrophoresis and alleles were scored by staining with ethidium bromide. Trisomy 21 was detected by the presence of three distinct alleles and transmission of alleles from parents to the offspring was determined in all but five families. Parental origin was found to be maternal in twenty three families and paternal in two families. The mean maternal and paternal ages of subjects were 25.1 ± 6.9 and 30.9 ± 6.9 years respectively. The results showed the use of molecular diagnosis for the allelic transmission from the parents to offspring and also emphasize the fact that the trisomy 21 was due to meiotic errors in maternal chromosomes.
Keywords: Down Syndrome, Trisomy 21, Parental Origin, PCR, D21S11 and D21S2055
Cite this paper: Tooba Altaf , Saba Irshad , "PCR Based Detection of Parental Origin of Extra Chromosome 21 in Children with Down Syndrome in Pakistan", Basic Sciences of Medicine , Vol. 1 No. 1, 2012, pp. 1-5. doi: 10.5923/j.medicine.20120101.01.
Article Outline
1. Introduction
- Down syndrome (DS) is the most commonly recognized human malformation complex and is the foremost known genetic cause of mental retardation. Down syndrome is a genetic disorder that results because of one cell has two 21 chromosomes instead of one, so the resulting fertilized egg has three sets of the 21st chromosomes. This abnormality is characterized by specific physical features and limited mental functions, along with several internal organ malformations, the established risk factors for Down syndrome include advancing maternal age and, family history of trisomy or relevant chromosomal rearrangements[1,2].Human Chromosome 21 was first mapped in May, 2000[3]. In general, this leads to an over expression of the genes[4]. Understanding the genes involved may help to target medical treatment to individuals with Down syndrome. It is estimated that chromosome 21 contains 200 to 250 genes. Research has identified a region of the chromosome that contains the main genes responsible for the pathogenesis ofDown syndrome, located proximal to 21q22.3. The search for major genes involved in Down syndrome characteristics is normally in the region 21q21–21q22.3[5].With the recombinant DNA technology a new set of tools became available to the study of origin and mechanisms of chromosomal abnormalities using DNA polymorphism analysis. In the beginning this kind of analysis used chromosome 21-specific DNA probes to detect restriction fragment length polymorphisms[6]. The development of the polymerase chain reaction (PCR) amplification technique[7] enabled the identification of novel and highly informative classes of DNA polymorphisms in the human genome, the so called microsatellites or simple sequence repeat (SSR) polymorphisms[8,9,10]. Especially the multi-allelic and easily typified microsatellites have contributed to mapping of the human genome[11,12] and to non-disjunction studies[13].The aim of the present study was to inverstigate the sensitivity and reliability of PCR diagnostic method for routine diagnosis of trisomy 21 and also for the determination of parental origin of nondisjoined chromosome.
2. Materials and Methods
2.1. Pateints & Methods
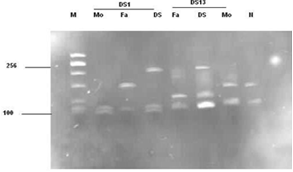
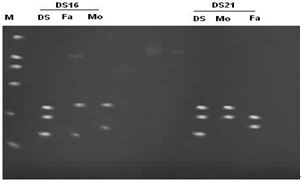
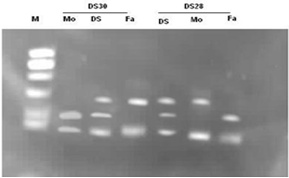
- Thirty families, DS1 to DS30, each with a clinically suspected Down syndrome child and a control normal individual was included in this research. After informed consent of Hospital authorities sampling was done in 7 months period (June 2007- December 2007). Blood samples were collected from paediatric genetics department of The Children’s Hospital & Institute of Child Health, (CH&ICH) Lahore. Blood samples were collected in heparin containing tubes for karyotyping and in sterile sodium-EDTA vacutainer tubes for molecular detection of Down syndrome. Complete physical examination of positive patients was performed and complete family history including maternal and paternal age at time of birth of trisomic band was recorded. Lymphocyte culture was set up by following the method[14,15]. Chromosome preparation was made from lymphocyte culture by conventional air drying method. For molecular analysis, genomic DNA was isolated from uncoagulated blood samples of trisomic probands and parents by using salting out procedure of Miller et al.[16]. DNA polymorphism was studied by using two tetranucleotide markers D21S11 and D21S2055 situated on chromosome 21 at 21q21 and 21q22 respectively. The sequence for the primers used was based on the published information as in[17]. The sequence of D21S11 forward primer is 5´ GTGAGTCAATTCCCCAAG 3´and of reverse primer5´ GTTGTATTAGTCAATGTTCTCC 3´. While the sequence of D21S2055 forward primer is 5´AACAGAAACCAA- TAGGCTATCTTATC 3´ and that of reverse primer 5´ TACAGTAAATCACTTGGTAGGAGA 3´The primers were diluted to 10µΜ. To perform PCR reaction, a master mix was prepared containing water, buffer (10X PCR buffer with (NH4)2SO4), dNTPs (Fermentas), reverse and forward primers and Taq DNA polymerase (Fermentas) in a single tube, which can then be aliquoted into individual tubes containing template DNA. Optimized concentration of MgCl2 (Fermentas) was then added. Optimized concentration of MgCl2 was then added. For the first primer, after initial denaturaton at 950C for 5 min., 30 cycles of PCR amplification were done at 950C for 30s; 570C for 30s; 720C for 30s and final extension for 9 min at 720C. For the second primer, after initial denaturation at 940C for 3 min, 30 cycles of PCR amplification were done at 950C for 30s ; 400C for 30s; 720C for 1 min and final extension for 3 min at 720C. PCR (polymerase chain reaction) reactions were carried out in a Appendorf Thermal Cycler. The amplicons were analyzed in 6% polyacrylamide gel and analysed in UV transilluminator after staining with ethidium bromide. The parental origin of supernumerary chromosome 21, and therefore, the parental origin of nondisjunction, was determined after scoring of the polymorphic alleles in the parents and the proband by following the method described by Ghosh and Dey[18].
3. Results
- The current research work involves PCR amplification of small tendem repeat (STR) markers located on human chromosome 21 and analysis by fluorescence based method by ethidium bromide to identify the presence of an additional allele on the third copy of the chromosome so thus confirming the cytogenetic analyses.
3.1. Cytogenetic Studies
- Analysis of chromosme and karyotype revealed a diploid count of 2n=47,+21 in probands of all thirty families. All the parents of the proband showed normal chromosome count and normal karyotype.
3.4. Molecular Studies of Molecular Studies
4. Discussion
- The current study of the karyotypes of thirty patients revealed that most prevalent type of chromosomal defect was trisomy. Translocation comes after trisomy and the Mosaicism was the least frequent. In India as in[20,21] investigated and found that free trisomy has higher ratio as compared to mosaicism and translocation. These findings are in close agreement with the present study. The trisomy genetic pattern was 86.6%, trisomy was 10% and mosaicism was 3.3% in current study. This is in agreement with[22]. Ahmad and coworkers[23] described the clinical features and cytogenetic analysis of patients with Down syndrome (DS) in Bahawalpur, Pakistan. Mongoloid slant, epicanthal folds, simian crease, flat nasal bridge, Retarded growth and development was observed in 90% cases of present research. The current research study showed that out of the total thirty patients, 53% were males and 46% were females. Devlin and his research group determined the accuracy of clinical diagnosis of Down syndrome. Simian crease, sandal gap, epicanthic folds, hypotonia, upslanting palpebral fissures, and protruding tongue were the most frequent characteristic features seen. These findings are in agreement with present study. So it shows that males have higher tendency than females to be affected with Down syndrome. The relation among demographic features, clinical features, and karyotype analyses of patients with Down syndrome (DS) was established as in[24]. In neonates, common features noted were mongoloid slant, ear abnormalities, flat facial profile, hypotonia[24,25,26]. On average, language and communication characteristics of individuals with Down syndrome (the most common genetic cause of intellectual disability) follow a consistent profile as in[27]. This is in direct relation with present study. There has not been any significant study describing prevalence of Down syndrome in PakistanThe analysis described here using the microsatellite markers/ short tendem repeats as in[28,29] D21S11 and D21S2055 showed that the origin of extra chromosome was maternal in about 92% of the cases of trisomy 21 and paternal in about 8%. These results contrast with those obtained in earlier molecular studies where paternal nondisjunction accounts for approximately 5%-6% of trisomy 21 cases only[30,19,31] . One study as in[32] showed the gender effects on the incidence of aneuploidy in mammalian germ cells. Maternal age remained the paramount aetiological factor associated with human aneuploidy. The majority of extra chromosomes in trisomic offspring appeared to be of maternal origin resulting from nondisjunction of homologous chromosomes during the first meiotic division. The current work is in accordance to their findings as origin of extra chromosome was maternal in about 92% of the cases of trisomy 21. The maternal age has deep impact on the incidence of Down’s syndrome in children. The mean maternal age for 30 Down syndrome families was 25.1 ± 6.9 years, not significantly different from the value of 30 ± 5.2 years reported by[19]. While in late thirties the risk of DS children increases. This is in close accordance as in[33,34]. One of them described the close association of maternal age with DS in UAE mothers. According to them the mean maternal age was 33.48+-8.08 years and 41.66% of the mothers were in the advanced maternal age group. The current study has elucidated that 36% mothers were in late thirties. Jyothi as in[35] determined the mean maternal age to be 30-34 years, which is close to the present work. In another study Jyothy explained that most of the translocation DS cases (n = 31) were born to younger mother's (< 25 years), when compared to pure trisomy 21 DS cases[36].Using molecular techniques, Chen and his group collected genomic DNA samples from 50 individuals diagnosed previously by karyotype as trisomy 21 and 40 children with severe mental retardation (IQ < 50) suspected of trisomy 21 were analyzed for 2 short tandem repeat loci on 21 chromosome, D21S1435 and D21S2055. Typing was carried out after polymerase chain reaction (PCR) and silver staining. The trisomy was identified by the number of alleles: 3 alleles bands whose density is same, two alleles bands with one obvious higher density compared to the other and one allele band whose density is three times than the normal control. STR marker D21S2055 was also used in the present research work to amplify the STR on chromosome 21. The allelic pattern was also same as in Chen’s work[37].A retrospective survey of all cases of Down syndrome was recorded between 1981 and 2000 to mothers resident in County Galway. The study compared the incidence of Down syndrome in both decades. Down syndrome in the second decade was directly related to the significant increase in the proportion of women in the 30 plus age group[38]. It is in accordance to present study as the incidence of having a Down syndrome baby is higher is the conceptions of late age of mothers.
5. Conclusions
- Down syndrome is the phenotypic consequence of trisomy 21 and is the most common genetically defined cause of intellectual disability. Currently, Down syndrome is one of the most common birth defects, affecting about one in every 750 live births. John Langdon Down first described this condition in the medical literature in 1866, documenting the various symptoms associated with the syndrome but failing to determine their cause. In fact, the cause of DS remained unknown for nearly 100 years following Down's work. Then, in the 1950s, researchers finally determined the source of DS: the presence of an extra copy of chromosome 21, a condition often referred to as trisomy 21. Fifteen percent of patients institutionalized for mental retardation have Down syndrome. The karyotype is usually performed by culturing leukocytes from a peripheral blood sample to determine the parental origin of the extra chromosome 21 in trisomy 21. Down syndrome is of importance for the understanding of the mechanism of nondisjunction and also for the elucidation of the maternal age effect. The recent research work was carried out in order to determine the parental origin of the extra chromosome 21 in trisomy 21 by molecular detection employing PCR. Since the discovery of trisomy 21, scientists have made great strides in Down syndrome research. For instance, researchers have identified a second (although less common) cause of the condition that is related to chromosomal translocation, and they have also determined the complete DNA sequence of chromosome 21. In addition, scientists have recently created mouse models of DS that possess chromosomal abnormalities similar to those of human patients. Through the use of these models, researchers hope not only to gain a better understanding of the specific genes that play the most significant role in DS, but also to eventually develop improved medical treatments for patients with this condition[38].
ACKNOWLEDGEMENTS
- We are very grateful to patients involved in this study and Dr. Saqib Mehmood, Head of the Department of Medical Genetics at CH&ICH, Lahore for providing access to affected patients and valuable suggestions during the study.
References
| [1] | Bell, J., 1991, The epidemiology of Down syndrome., Med. J. Aust., 155,115–117 |
| [2] | Leck, I., 1994, Structural birth defects. The Epidemiology of Childhood Disorders., 66–117 |
| [3] | Hattori, M., 2000, The DNA sequence of human chromosome 21., Nature, 405, 311-319 |
| [4] | Rong, M.X., Wang, E.L., Spitznagel, L.P., Frelin, J.C., Ting, H. D., J, Kim, I., Ruczinski, T.J., Downey, J.P., 2005, Primary and secondary transcriptional effects in the developing human Down syndrome brain and heart., Genome Biology, 6 (13), 107 |
| [5] | Rahmani, Z., Louis, J., Blouin, Nicole, Créau-Goldberg, Paul, C., Watkins, Jean-François, Mattei, Marc, Poissonnier, Marguerite, Prieur, Zoubida, Chettouh, Annie, Nicole, Alain, Aurias, Pierre-Marie Sinet, Jean-Maurice Delabar., 2005, Down syndrome critical region around D21S55 on proximal 21q22.3., Am. J. Md. Gen., 37, 98-103 |
| [6] | Davies, K.E., Harper, K., Bonthron, D., 1984, Use of a chromosome 21 cloned DNA probe for the analysis of non-disjunction in Down syndrome., Hum. Genet., 66, 54–56 |
| [7] | Saiki, R.K., Gelfand, D.H., Stoffel, S. 1988, Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase., Science, 239, 487–491 |
| [8] | Weber, J.L., and May, P.E., 1989, Abundant class of human DNA polymorphisms which can be typed using the polymerase chain reaction., Am. J. Hum. Genet., 44, 388–396 |
| [9] | Litt, M., and Luty, J.A., 1989, A hypervariable microsatellite revealed by in vitro amplification of a dinucleotide repeat within the cardiac muscle actin gene., Am. J. Hum. Genet., 44, 397–401 |
| [10] | Economou, E.P., Bergen, A.W., Warren, A.C., 1990, The polydeoxyadenylate tract of Alu repetitive elements is polymorphic in the human genome., Proc. Natl. Acad. Sci., 87, 2951–2954 |
| [11] | NIH/CEPH Collaborative Mapping Group, 1992, A comprehensive genetic linkage map of the human genome., Science, 258, 67–86 |
| [12] | Weissenbach, J., Gyapay, G., Dib, C., 1992, A second -generation linkage map of the human genome., Nature, 359, 794–801. |
| [13] | Petersen, M.B., Schinzel, A.A., Binkert, F., 1991, Use of short sequence repeat DNA polymorphisms after PCR amplification to detect the parental origin of the additional chromosome 21 in Down syndrome., Am. J. Hum. Genet., 48, 65–71 |
| [14] | Worton, R.G., and Duff, C., 1979, Karyotyping. Method., Enzymol., 58, 322-344 |
| [15] | Roong, D., and E., Czepulkowski. B, H., 1992, Human Cytogenetics., Volume 1 Constitutional analysis, A practical approach, Oxford Press |
| [16] | Miller, S.A., Dykes, D.D., Polesky, H,F., 1988, A simple salting out procedure for extracting DNA from human nucleated cells., Nucl.Acids, Res., 16, 1215 |
| [17] | Ghosh, S., and Dey, S.K., 2005, PCR based detection of parental origin of extra chromosome 21 in Down Syndrome. Int. J. Hum. Genet., 5(3), 183-186 |
| [18] | Ghosh, S. and Dey, S.K., 2003, DNA diagnosis of Down syndrome using polymerase chain reaction and polymorphic microsatellite markers., Int. J. Hum. Genet., 3(1), 17-20 |
| [19] | Sherman, S.L., Takaesu, N., Freeman, S.B., 1991, Trisomy 21: association between reduced recombination and nondisjunction., Am. J. Hum. Genet., 49,608–620 |
| [20] | Malini, S.S., and Ramachandra, N.B., 2006, Influence of advanced age of maternal grandmothers on Down syndrome., B.M.C. Med. Genet., 14, 4-7 |
| [21] | Jain, S., Agarwal, S., Panigrahi, I., Tamhankar, P., Phadke, S., 2010, Diagnosis of Down syndrome and detection of origin of nondisjunction by short tandem repeat analysis.,Genet.Test.Mol.Biomarkers.,(4),489-91 |
| [22] | Catovic, A., and Kendic, S., 2005, Cytogenetic findings at Down syndrome and their correlation with clinical findings., Bosn. J. Basic, Med. Sci., 5(4),61-7 |
| [23] | Ahmed, I., Ghafoor, T., Samore, N.A., and Chattha, M.N., 2005, Down syndrome: clinical and cytogenetic analysis., J. Coll. Physicians, Surg. Pak., 15(7), 426-429 |
| [24] | Bhattacharyya, R., Sanyal, D., Roy, K., and Bhattacharyya, S., 2010, Correlation between physical anomaly and behavioral abnormalities in Down syndrome., J. Pediatr. Neurosci., 5(2),105–110 |
| [25] | Sureshbabu, R., Kumari, R., Ranugha, S., Sathyamoorthy, R., Udayashankar, C., Oudeacoumar, P., 2011, Phenotypic and dermatological manifestations in Down Syndrome., Dermatol., Online, J.,17(2),3 |
| [26] | Lu, Y.P., Cheng, J., Jiang, S.F., Zhang, L.W., Gao, Z.Y., Han, B., Yuan, H.J., Li, Y.L., 2010, Development of multiple quantitative fluorescent PCR for rapid diagnosis of common aneuploidy and it's clinical application., Yi Chuan. 32(11),1141-6 |
| [27] | Martin, G.E., Klusek, J., Estigarribia, B., Roberts, J.E., 2009, Language Characteristics of Individuals with Down Syndrome., Top Lang Disord. 29(2),112-132 |
| [28] | Crkvenac-Gornik, K., Grubić, Z., Stingl, K., Muzinić, D., Brkljacić-Kerhin, V., Begović, D., 2007, Rapid prenatal diagnosis of numerical aberrations of chromosome 21 and 18 by PCR-STR method., Coll. Antropol., 31(3),859-62 |
| [29] | Kava, M.P., Tullu, M.S., Muranjan, M.N., Girisha, K.M. 2004, Down syndrome: clinical profile from India., Arch. Med. Res., 35(1), 31-35 |
| [30] | Antonarakis, S.E., Lewis, J.G., Adelsberger, P.A., 1991, Parental origin of the extra chromosome in trisomy 21 as indicated by analysis of DNA polymorphisms., N. Engl. J. Med., 324, 872–876 |
| [31] | Ko, T.M., Hwa, H.I., Tseng, L.H., Lin, Y.W., Cheung, Y.P., 1998, Fluorescence microsatellite analysis to study the parental origin of the supernumerary chromosome in Down’s syndrome., Int, J, Gynaecol, Obstet., 61(2), 149-153 |
| [32] | Langlois, S., Duncan, A., 2011, Use of a DNA Method, QF-PCR, in the Prenatal Diagnosis of Fetal Aneuploidies., J. Obstet. Gynaecol. Can., 33(9),955-60 |
| [33] | Pacchierotti, F., Adler, I.D., Eichenlaub-Ritter, U., Mailhes, J.B., 2007, Gender effects on the incidence of aneuploidy in mammalian germ cells., Environ. Res., 104(1),46-69 |
| [34] | Murthy, S.K., Malhotra, A.K., Mani, S., Shara, M.E., Al-Rowaished, E.E., Naveed, S., Alkhayat, A., Alali, M.T., 2007, Incidence of Down syndrome in Dubai., UAE. Med. Princ. Pract., 16(1),25-28 |
| [35] | Jyothy, A., Kumar, K.S., Mallikarjuna, G.N., Babu, R.V., Uma, D. B., Sujatha, M., Reddy, P.P., 2001, Parental age and the origin of extra chromosome 21 in Down syndrome., J. Hum. Genet., 46(6),347-350 |
| [36] | Jyothy, A., Rao, G.N., Kumar. K.S., Rao, V.B., Uma, D. B., Reddy, P,P., 2002, Translocation Down syndrome., Indian, J. Med. Sci., 56(5), 225-229 |
| [37] | Chen, H., Xin, J., Li, N., Liang, W., Liao, M., Chen, G., Wu, K., Zhang, L., 2002, A method for rapid and early diagnosis of trisomy 21 using molecular techniques., Hua, Xi, Yi, Ke, Da, Xue, Xue, Bao., 33(1),125-8 |
| [38] | O'nuallain, S., Flanagan, O., Raffat, I., Avalos, G., Dineen, B., 2007, The prevalence of Down syndrome in County Galway., Ir. Med. J., 101(1), 329-331 |
| [39] | O'Connor, C., 2008, Trisomy 21 causes Down syndrome. , Nature Education 1(1) |
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-Text HTML
Full-Text HTML