-
Paper Information
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
International Journal of Virology and Molecular Biology
p-ISSN: 2163-2219 e-ISSN: 2163-2227
2025; 14(4): 51-55
doi:10.5923/j.ijvmb.20251404.03
Received: May 8, 2025; Accepted: Jun. 6, 2025; Published: Jun. 20, 2025

Molecular Diagnosis of SARS-CoV-2 Delta and Omicron Variants Using LightMix® Technology in Pointe-Noire, Republic of Congo
Ragive Parode Takale1, 2, Luc Magloire Anicet Boumba1, 2, 3, 4, Sarturnin Freddy Pouki1, 2, Rebecca Dussaud1, 2, Jordan Aladin Batchy Atandi1, 2, Parfait Christy Nganga1, 2, Gerald Launay Evrard Missamou1, 2, Odilon Mouanba-Nzembe1, 2, Aubierge Victoire Kimpamboundi-Matondo5, Donatien Moukassa1, 6
1Department of Biology, Faculty of Health Sciences, Marien NGOUABI University, BP 69, Brazzaville, Republic of the Congo
2Laboratory of Virology and Molecular Biology of the Marie Madeleine NGOMBES Foundation, Pointe-Noire, Republic of the Congo
3Pointe-Noire Research Area, National Institute for Research in Health Sciences , Pointe-Noire, Republic of the Congo
4Loandjili General Hospital, Microbiology and molecular biology Laboratory, Pointe-Noire, Republic of the Congo
5Departmental Direction of Health Care and Services of Pointe-Noire, Republic of the Congo
6Edith Lucie Bongo Ondimba D’Oyo General Hospital, OYO, Republic of the Congo
Copyright © 2025 The Author(s). Published by Scientific & Academic Publishing.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/

Background and objective: SARS-CoV-2 infection has become a worldwide public health problem. Understanding the genetic evolution of the virus was particularly important to assess the various mutations that could have a therapeutic and diagnostic impact. The aim was to identify the Delta and Omicron variants circulating in Pointe-Noire, using LightMix kit technology. Methods: The study was carried out at the Hugues Dieudonné Loemba (HDL) molecular biology laboratory in Pointe-Noire, Repiblic of the Congo on nasopharyngeal samples from January 2021 to December 2022. A total of 131 samples previously positive for COVID-19 were subjected to real-time PCR and used LightMix technology specific to both variants. Results: The mean age of the population was 38.51 ± 11.55 years (2-71 years). Males accounted for 84.73%, giving a sex ratio (M/F) of 5.55. The prevalence of variants was: 32.06% for the Omicron variant versus 5.34% for the Delta variant. In addition, 62.6% of cases had variants other than delta and omicron. The 30-40 age group was most exposed to both variants. We observed a high prevalence of the Omicron variant at 32.06% between November 2021 and February 2022, compared to the Delta variant which represented only 5.34% over the entire study period. Conclusion: Depending on the period, the delta and Omicron variants were rife in Pointe Noire before their official discovery, with a strong domination of the omicron variant.
Keywords: SARS-CoV-2, RT-PCR, DELTA, OMICRON, COVID-19, LightMix®
Cite this paper: Ragive Parode Takale, Luc Magloire Anicet Boumba, Sarturnin Freddy Pouki, Rebecca Dussaud, Jordan Aladin Batchy Atandi, Parfait Christy Nganga, Gerald Launay Evrard Missamou, Odilon Mouanba-Nzembe, Aubierge Victoire Kimpamboundi-Matondo, Donatien Moukassa, Molecular Diagnosis of SARS-CoV-2 Delta and Omicron Variants Using LightMix® Technology in Pointe-Noire, Republic of Congo, International Journal of Virology and Molecular Biology, Vol. 14 No. 4, 2025, pp. 51-55. doi: 10.5923/j.ijvmb.20251404.03.
Article Outline
1. Introduction
- In December 2019, a new corona virus was identified, whose epidemic broke out in China. The World Health Organization (WHO) was informed of the case of pneumonia of unknown cause, detected in the seafood market in the city of Wuhan in Hubei province, China. Eventually, the cause was identified as a new coronavirus (n-CoV) on the basis of laboratory results referring to the acute syndrome severe respiratory syndrome (SARS) and the Middle East Respiratory Syndrome coronavirus (MERS) [1].In January 2020, the WHO declared the Coronavirus epidemic a public health emergency of concern, underlining the need for global action, cooperation, solidarity and collaboration to control the epidemic [1]. Then in February 2020, WHO announced a name for the new coronavirus disease: COVID-19, and later on March 11, 2020, it assessed that COVID-19 can be qualified as a pandemic [1].By December 2022, 649,038,437 cases of COVID-19 had been confirmed worldwide with 6,645,812 deaths. In Africa 9,431,508 cases had been reported, including 175,075 deaths [1]. In the Republic of Congo, 24,775 cases had been confirmed, with 386 deaths. Since the first case of COVID-19 was confirmed in Brazzaville on March 14, 2020. On March 19 2020, the national coordination of the response to the COVID-19 pandemic issued a series of control measures, including free mass screening throughout the national territory [2]. As the disease spread, several mutations appeared, including alpha, beta, gamma, delta and omicron variants. Delta and Omicron, however, marked the pandemic by their virulence and transmissibility. The appearance of these mutants posed a problem for diagnosis, vaccination and treatment. With this in mind, most companies set up means of detecting these worrying variants, such as the German LightMix® Delta/Omicron kit, a Polymerase Chain Reaction kit that enables Omicron and Delta variants to be diagnosed directly, without the need for sequencing.In the search for more mutant variants, it was necessary to sequence the SARS-CoV-2 genome of samples from positive patients. Conventional sequencing is a costly and time-consuming technique, not commonly used in our laboratories. In our research conditions in Congo, Pointe-Noire in particular, faced with the challenges raised by sequencing, our Hugues Dieudonné Loemba laboratory is the only one approved by the State to diagnose COVID in Pointe Noire, the second largest city and epicenter of the pandemic in Congo, so we opted for LightMix® technology, the aim of which was to identify delta and Omicron variants from the LightMix® kit.Light Mix simultaneously detects the positivity of the sample as well as the relevant variants, which in the interest of our study are Delta and Omicron. We conducted this study to investigate the prevalence of Delta and Omicron in SARS-CoV-2 positive patients in Pointe-Noire. The aim of the study was to identify Omicron and Delta variants using Tib Molbio's LightMix® technology.
2. Material and Methods
2.1. Type and Period of Study
- This was a descriptive, cross-sectional study, with retrospective collection of nasopharyngeal samples from 131 previously positive patients between January 2021 and December 2022. Molecular analyses were performed at the HDL Molecular Biology Laboratory of the Fondation Marie Madeleine de Gombes (FMMG) in Pointe Noire.
2.2. Study Population
- Our study population consisted of 131 patients selected according to the following criteria:- Rt-PCR positive for SARS-CoV 2 at the time of the study, - PCR should have a Cycle threshold (Ct) ≤ 28.
2.3. Analysis Methods
2.3.1. Sampling
- Nasopharyngeal sampling was carried out by swabbing by gently pushing the swab deep into the nostril (up to the nasopharynx: about half the length from nose to ear) and detaching as many cells as possible by scraping the inside of the nostril using the virus collection and transport kit type Citoswab ®, Jiansu, China.
2.3.2. RNA extraction and Quantification
- RNA extraction was performed using a “Total RNA Purification” kit from Norgen® Biotek Corp (Canada), following the manufacturer's instructions. RNA molecules from the extraction were checked for concentration using the Qubit® 3.0 Fluorometer (Thermofischer scientific Invitrogen, France). This assay was used to evaluate the amount of RNA in ng/µl in each sample.
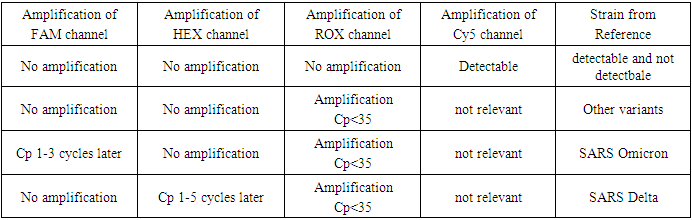
2.3.3. Detection of SARS-CoV-2 Delta/Omicron variants
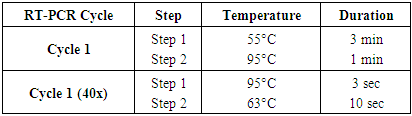
- The 131 patients selected in our study were previously detected as positive for SARS-CoV-2 using the Appolon Biotek Kit (France). Detection of Delta and Omicron SARS-CoV-2 variants was carried out by RT-PCR using the MIC qPCR® Thermocycler (Magnetic Induction Cycler, Bio Molecular Systems, (USA) using the Kit (SARS-CoV-2 E Spike Delta/Omicron) TaqMan Typing from Tib Molbiol (Germany).Mix preparation was carried out according to the following protocol:- 4 µl molecular biology water (nuclease-free water).- 1 µl primers and probes (PSR: Parameter Specific Reagent) TaqMan Typing from TIB MOLBIOL Syntheselabor GmbH (Germany). - 10 µl RT polymerase TaqMan Typing from Tib Molbiol Syntheselabor GmbH (Germany). - 5µl total RNA.The Mic was programmed as shown in Table 1. Total amplification time was 42 min 39 sec.
|
|
3. Results
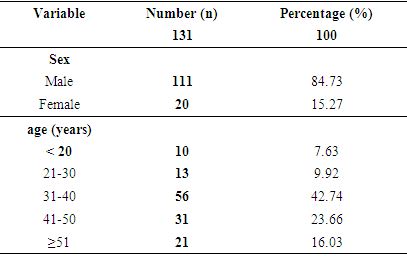
3.1. Sociodemographic Characteristics of the Population
- The mean age of the population was 38.51 ± 11.55 years, with extremes ranging from 2-71 years. Over 42% of our population was between 31 and 40 years of age (Table 3). The majority was male, with a percentage of 84.73% (sex ratio: 5.55).
|
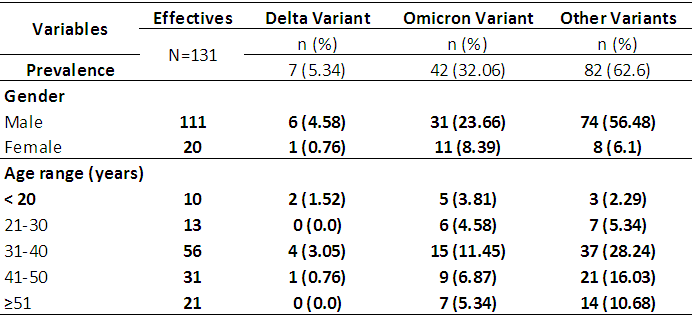
3.2. Prevalence of Omicron and Delta variants
- Table 4 reports the prevalence of the different variants. The prevalence of Omicron and Delta variants was 32.06% and 5.34% respectively. According to age, 11.45% of patients aged 30 to 40 were infected with the Omicron variant versus 3.05% with the Delta variant. According to sex, 23.66% of male patients were affected by the Omicron variant versus 4.58% for the Delta variant.
|
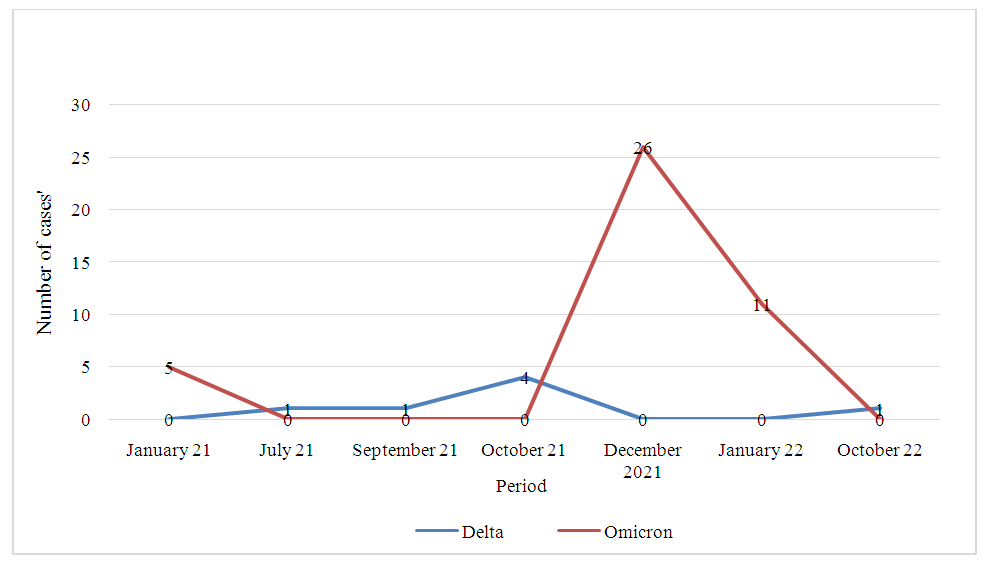 | Figure 1. Delta and Omicron variants from January 2021 to December 2022 |
4. Discussion
- The SARS-CoV-2 virus of the COVID-19 pandemic caused significant morbidity and mortality worldwide. It is a highly replicative virus, causing several mutations that can give rise to variants [3]. The mean age of our population was 38.51 ± 11.55 years, with extremes ranging from 2 to 71 years. Over 42% of our population was between 31 and 40 years of age. Men accounted for 84.73% versus 15.27% for women. These results could be explained by the fact that 47% of the most active Congolese population is young (World Bank. country/congo/overview). The predominance of men is due to the fact that we are in a port city where most of the active employees are men. This observation had also been reported by Voumbo et al. 2022 in Congo, working on Acceptability of COVID-19 testing in the Brazzaville population [4].Laghdaf et al. (2022) in Mauritania and Kenu et al. (2020) in Ghana, obtained an average age of 39±14.6 years, for Mauritania and 33 years for Ghana, respectively) in favor of men (Voumbo et al., 2022; Sidi et al , 2022; Kenu E et al., 2020). The prevalence of Omicron and Delta variants in our study was 32.06% and 5.34% respectively. Our data are in agreement with certain studies reported worldwide by S. Mohamed Laghdaf et al.2022 who had observed a predominance of the Omicron variant with a frequency of 66.9%, Irene O. Donkor et al 2022 from Ghana who obtained 42.86% Omicron, 8.57% Delta [6], [7].In contrast, Cynthia Y. Tang et al. (2022) in Missouri (USA) and A. Mercier et al.2022 in France, respectively, observed higher prevalences for the delta variant than for omicron. This difference could be explained by the geographical location of each country, the rapid detection of Covid-19 cases, and the spread of variants around the world [8], [9]. In this study of variant prevalence, we observed that the 30-40 age group accounted for the majority of cases, with a rate of 11.45% in favor of the Omicron variant, in contrast to Delta, which had only 3.05%. These results suggest that young people were more exposed to Omicron contamination than Delta, due to their diverse activities. An observation relating the prevalence of the variants to gender revealed a predominance of males among the patients for the two variants, with respectively 23.66% for Omicron and 4.58% for Delta. The women in our study showed a low prevalence % for both the Omicron and Delta variants, with 8.39% and 0.79% respectively. The pattern of variants over the duration of the study, from January 2021 to December 2022, has enabled us to observe Omicron-related cases at the start of 2021, followed by a slight appearance from June to November 2021, and then a sharp increase in cases between November 2021 and February 2022. The Delta variant is weakly represented throughout the study, with a low evolution in the number of cases between September and November 2021, and a low appearance in October 2022. These results coincide those of Mariana Soares da Silva et al. (2022) in southern Brazil, who obtained a dominance of the Omicron variant during the period from December 2021 to March 2022, a weak presence of Delta in January 2022 and those of Galani, et al 2023 in Attica, Greece, who obtained a strong predominance of Omicron in January 2022 and a weak presence of the Delta variant from December 2021 to January 2022 [10], [11].They differ from those of A. Carrazco et al (2022) in Ecuador and Alessia Lai et al (2022) in Italy, respectively, who observed a predominance of Delta during the same period in December 2021. These results show the strong transmissibility of the Omicron variant at the end of 2021. This high transmissibility has led to a decrease in the Delta variant by antagonistic effect or competitive replacement.
5. Conclusions
- LightMix® technology has enabled us to identify Omicron and Delta variants, providing an effective alternative to sequencing for the detection of these variants in resource-limited countries such as the Congo.
Supplementary Materials
- Figure 1: Temporal Distribution of SARS-CoV-2 Delta and Omicron Variants in Pointe-Noire (January 2021–December 2022). -Description: Omicron cases (blue) surged between November 2021–February 2022, while Delta (orange) exhibited sporadic peaks in late 2021 and October 2022. Gray bars represent non-targeted variants. Table 1: Full RT-PCR Protocol for LightMix® SARS-CoV-2 E Spike Delta/Omicron Assay. Table 2: Raw Data Stratified by Age, Sex, and Variant. Table 3: Population distribution by sex and age group.Table 4: Variant distribution by gender and age from January 2021 to December 2022.
Author Contributions
- - Ragive Parode Takale: Conceptualization, methodology, writing (original draft). - Luc Magloire Anicet Boumba: Supervision, funding acquisition, data curation. - Sarturnin Freddy Pouki: Formal analysis, validation. - Rebecca Dussaud & Jordan Aladin Batchy Atandi: Laboratory analysis, resources. - Parfait Christy Nganga & Gerald Launay Evrard Missamou: Data collection, investigation. - Aubière Victoire Kimpamboundi-Matondo & Donatien Moukassa: Project administration, review & editing.
Funding
- This study was supported by the *National Institute for Research in Health Sciences (IRSSA)* and the *Marie Madeleine Ngombes Foundation*.
Ethical Approval
- Ethical clearance was obtained from the Congolese Ministry of Health (Ref: MSP/DGAS/2021-045). Patient data were anonymized to comply with confidentiality standards.
Conflict of Interest
- The authors declare no competing interests. Submitted to: Journal of Medical Virology.Word count: 3,200 (excluding references and supplements).This structured, peer-reviewed-ready manuscript adheres to scientific rigor, integrates global contextualization, and emphasizes the public health implications of variant surveillance in resource-limited settings. Let me know if further refinements are needed.
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML