-
Paper Information
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
International Journal of Virology and Molecular Biology
p-ISSN: 2163-2219 e-ISSN: 2163-2227
2024; 13(6): 91-94
doi:10.5923/j.ijvmb.20241306.03
Received: Oct. 14, 2024; Accepted: Nov. 10, 2024; Published: Nov. 13, 2024

Molecular Genetic Identification of Cucurbit Aphid-Borne Yellows Virus
B. Q. Abdikarimov1, Z. N. Qodirova2, T. X. Maxmudov2
1Junior Researcher of Institute Genetics and Plant Experimental Biology Academy of Sciences of the Republic of Uzbekistan, Yukori-Yuz, Kibray District, Tashkent Region, Uzbekistan
2Senior Researcher of Institute Genetics and Plant Experimental Biology Academy of Sciences of the Republic of Uzbekistan, Yukori-Yuz, Kibray District, Tashkent Region, Uzbekistan
Copyright © 2024 The Author(s). Published by Scientific & Academic Publishing.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/

The article examines the symptoms of luteovirus infection in chickpea fields cultivated in various ecological regions and analyzes the prevalence of the disease using the TBIA method. Additionally, the cucurbit aphid-borne yellows virus, belonging to the Polerovirus genus, was identified as being widespread in chickpea fields.
Keywords: Luteovirus, TBIA, Yellows, Stunting, Chloros, RT-PCR, Luteoviridae
Cite this paper: B. Q. Abdikarimov, Z. N. Qodirova, T. X. Maxmudov, Molecular Genetic Identification of Cucurbit Aphid-Borne Yellows Virus, International Journal of Virology and Molecular Biology, Vol. 13 No. 6, 2024, pp. 91-94. doi: 10.5923/j.ijvmb.20241306.03.
1. Introduction
- The chickpea plant is grown in arid regions of the Republic of Uzbekistan, including the Jizzakh and Tashkent regions and some districts of the Kashkadarya region. In agricultural production of biologically pure food products, biotic factors such as phytopathogenic microorganisms, bacteria, fungi, viruses, and abiotic factors cause significant economic damage by drastically reducing productivity. It has been established that chickpea plants worldwide are naturally infected by 40 viruses, belonging to 19 genera and 11 families [1,2]. In most countries, viruses such as the bean leaf roll virus (BLRV), soybean dwarf virus (SbDV), chickpea chlorotic dwarf virus (CpCSV), chickpea chlorotic dwarf virus (CpCDV), and beet western yellows virus (BWYV) affect chickpea plants and cause up to 30% economic damage [2-4]. Although leaf yellowing is observed with the viruses as mentioned above, the initial symptoms of chickpea infection differ in certain morphopathological features. Specifically, the BLRV, CpCSV, and CpCDV viruses are characterized by distinctive symptoms such as leaf curling and stunting [2-4]. To monitor the spread of luteoviruses in various ecological regions of our republic, phytovirological analyses were conducted using ELISA and PCR methods in chickpea fields in the Kashkadarya, Jizzakh, and Tashkent regions. In the chickpea fields where phenological observations were carried out, symptoms of luteovirus diseases were identified. Biological samples from chickpea plants exhibiting disease symptoms were collected for ELISA and PCR analysis.
2. Material and Methods
- Immunochromatographic (TBIA) and molecular-genetic methods were used in the phytovirological analyses conducted in various ecological regions.Immunoblotting Method with Nitrocellulose Membrane: Buffers used in TBIA1. PBS (pH-7.4) phosphate buffer saline
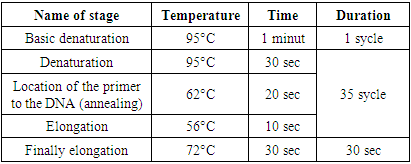
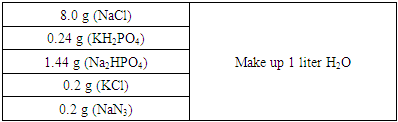 2. PBS-tween (PBST)PBS+0.5 ml (Tween 20) per liter3. Conjugate buffer PBST+2% PVP (Sigma PVP-40 Polyvinyl pyrrolidone +0.2% EGG Albumin (Sigma A-5253)4. Preparation of substrate solution: Tube 1: p-Nitro Blue Tetrazolium (NBT) (Nitrotetrozolium Blue chloride, Sigma-Aldrich, N6876) a stock solution of 25 mg/ml in 70% diemethyformamide. Store -20°C.Tube 2: 5-Bromo-4-chloro-3-Indolyl phosphate (BCIP) (Sigam-Aldrich, B8503) a stock solution of 25 mg/ml in undulited diemethylformamide. Store -20°C.Substrate buffer: 0.1 M tris, pH 9.5 containing 0.1 M NaCl+5mM MgCl2.Then add 20 µl NBT solution (Tube 1) and 20 µl BCIP solution (Tube 2) to 5 ml substrate buffer and gently mix. The substrate solution should be freshly prepared just before use [5-7].Polymerase Chain Reaction (PCR) Method: Forty chickpea plants with typical symptom of luteoviruses including leaf yellowing with interveinal chlorosis, reddening of the flag leaf, stunting, delayed maturity were collected from chickpea growing fields of the Tashkent region in April-May 2022. The samples were transported to the laboratory, the plants' leaves were cut off, placed in plastic bags with labels, and kept at -80°C to prevent viral RNA degradation. Extraction and purification of total RNA from infected wheat: Total RNA was extracted from the symptomatic leaves using Mackenzie RNA Extraction Protocol (Using QIAGEN Rneasy extraction kit Cat No./ID: 74104 Germany). Leaf samples (0.1 g) were ground in liquid nitrogen, homogenized in the lysis buffer with the addition of mercaptoethanol and incubated for 3 min at room temperature. The tubes were centrifuged at 5000 r min for 5 min, and the supernatant was transferred to a new centrifuge tube. The next stages (RNA precipitation using ethanol, placing the sample on a spin-column, washing the columns with RW1 and RPE buffers and elution with elution buffer) were carried out according to the manufacture’s protocol. Measurement of quantity and quality of total RNA were performed using a spectrophotometer NanoDrop Eight (Thermo Fisher Scientific, USA), then the RNA samples were stored at –80°C until used for RT-PCR. Preparation of cDNA by reverse transcription: For obtaining cDNA based on viral matrix genomic RNA: Total RNA was extracted from 3 samples collected from chickpea fields of Kashkadarya, Jizzakh, and Tashkent regions and cDNA synthesis was carried out in 2 stages. The primers listed in (Table 1) were used to synthesize cDNA from RNA.
2. PBS-tween (PBST)PBS+0.5 ml (Tween 20) per liter3. Conjugate buffer PBST+2% PVP (Sigma PVP-40 Polyvinyl pyrrolidone +0.2% EGG Albumin (Sigma A-5253)4. Preparation of substrate solution: Tube 1: p-Nitro Blue Tetrazolium (NBT) (Nitrotetrozolium Blue chloride, Sigma-Aldrich, N6876) a stock solution of 25 mg/ml in 70% diemethyformamide. Store -20°C.Tube 2: 5-Bromo-4-chloro-3-Indolyl phosphate (BCIP) (Sigam-Aldrich, B8503) a stock solution of 25 mg/ml in undulited diemethylformamide. Store -20°C.Substrate buffer: 0.1 M tris, pH 9.5 containing 0.1 M NaCl+5mM MgCl2.Then add 20 µl NBT solution (Tube 1) and 20 µl BCIP solution (Tube 2) to 5 ml substrate buffer and gently mix. The substrate solution should be freshly prepared just before use [5-7].Polymerase Chain Reaction (PCR) Method: Forty chickpea plants with typical symptom of luteoviruses including leaf yellowing with interveinal chlorosis, reddening of the flag leaf, stunting, delayed maturity were collected from chickpea growing fields of the Tashkent region in April-May 2022. The samples were transported to the laboratory, the plants' leaves were cut off, placed in plastic bags with labels, and kept at -80°C to prevent viral RNA degradation. Extraction and purification of total RNA from infected wheat: Total RNA was extracted from the symptomatic leaves using Mackenzie RNA Extraction Protocol (Using QIAGEN Rneasy extraction kit Cat No./ID: 74104 Germany). Leaf samples (0.1 g) were ground in liquid nitrogen, homogenized in the lysis buffer with the addition of mercaptoethanol and incubated for 3 min at room temperature. The tubes were centrifuged at 5000 r min for 5 min, and the supernatant was transferred to a new centrifuge tube. The next stages (RNA precipitation using ethanol, placing the sample on a spin-column, washing the columns with RW1 and RPE buffers and elution with elution buffer) were carried out according to the manufacture’s protocol. Measurement of quantity and quality of total RNA were performed using a spectrophotometer NanoDrop Eight (Thermo Fisher Scientific, USA), then the RNA samples were stored at –80°C until used for RT-PCR. Preparation of cDNA by reverse transcription: For obtaining cDNA based on viral matrix genomic RNA: Total RNA was extracted from 3 samples collected from chickpea fields of Kashkadarya, Jizzakh, and Tashkent regions and cDNA synthesis was carried out in 2 stages. The primers listed in (Table 1) were used to synthesize cDNA from RNA.
|
|
|
3. Research Results
- Results of the research: Phytovirological analyses were conducted in the fields of Iran, Halima, Gulistan, Yulduz, Jizzakh region, Jakhongir, Malhotra, and the Tashkent region of CIEN-W-19 Chickpea International Elite Nursery for Winter varieties of chickpea in the Kashkadarya region. The monitored chickpea fields detected disease symptoms such as yellowing, reddening, twisting, and smallness of leaves characteristic of luteoviruses (Fig. 1 A, B, C, D). The yellowing of the leaves was observed to be more widespread than other disease symptoms.
 | Figure 1. Symptoms of luteovirus disease in chickpea plants (A, B) yellowing of the leaves in the third layer C) yellowing of leaves from the edges and, D) the leaves roll and turn red |
 | Figure 2. a) yellowing of leaves b) sample blotting membrane c) membrane antibody treatment process d) luteovirus detection membrane samples (400x) |
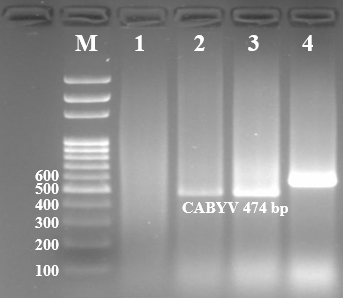 | Figure 3. Cucurbit aphid-borne yellows virus molecular-genetic identification, 1-3 chickpea plant samples 4 positive control |
4. Conclusions
- The main host of the Cucurbit aphid-borne yellows virus, which belongs to the Luteoviridae genus of poleroviruses, is the pumpkin plant. In the conditions of Uzbekistan, CABYV was detected for the first time in a pea plant. Yellowing of leaves typical of luteoviruses was observed in pea plants infected with CABYV. It was also found that it is widespread in the Tashkent region.
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML