-
Paper Information
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
International Journal of Virology and Molecular Biology
p-ISSN: 2163-2219 e-ISSN: 2163-2227
2024; 13(6): 81-86
doi:10.5923/j.ijvmb.20241306.01
Received: Sep. 28, 2024; Accepted: Oct. 22, 2024; Published: Oct. 30, 2024

Morphological and Molecular Identification of New Findings of Green Algae (Scenedesmaceae, Chlorophyta) from River Naryn, Uzbekistan
N. B. Ikramov
PhD student, Namangan State University, Namangan, Uzbekistan
Correspondence to: N. B. Ikramov, PhD student, Namangan State University, Namangan, Uzbekistan.
Copyright © 2024 The Author(s). Published by Scientific & Academic Publishing.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/

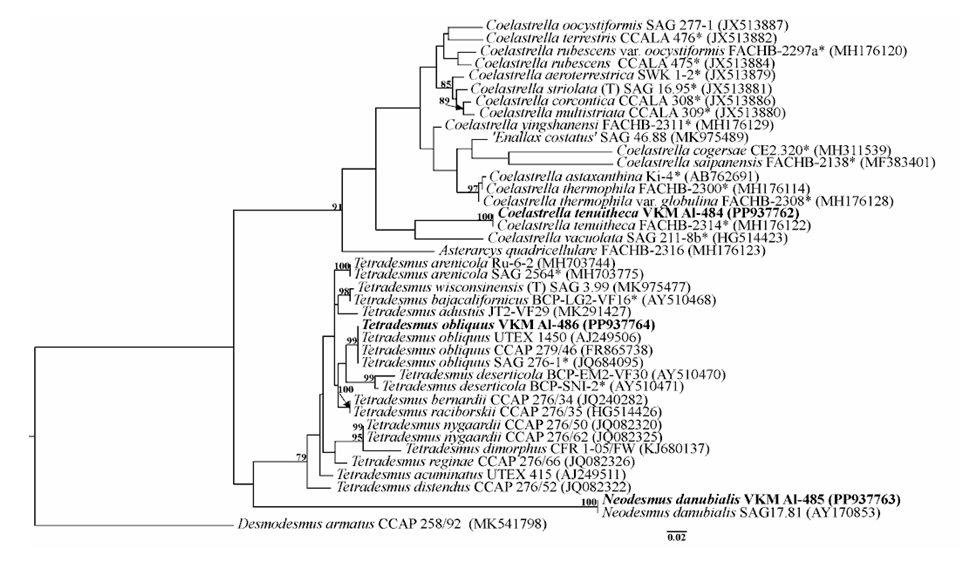
For the first time, the presence of green algal species Coelastrella tenuitheca, Neodesmus danubialis and Tetradesmus obliquus (Scenedesmaceae, Chlorophyta) was identified in the phytoplankton of the Naryn River (Uzbekistan) based on morphological and molecular analysis. Some strains showed discrepancies with their primary morphological diagnoses. This study confirms that ITS2 can be successfully used as a DNA barcode for future monitoring of freshwater ecosystems.
Keywords: Morphological identification, New findings, Green algae, Molecular identification, Scenedesmaceae, Primary morphological diagnoses, Chlorophyta, Naryn River, ITS2, Phytoplankton, DNA barcode, Freshwater ecosystems
Cite this paper: N. B. Ikramov, Morphological and Molecular Identification of New Findings of Green Algae (Scenedesmaceae, Chlorophyta) from River Naryn, Uzbekistan, International Journal of Virology and Molecular Biology, Vol. 13 No. 6, 2024, pp. 81-86. doi: 10.5923/j.ijvmb.20241306.01.
Article Outline
1. Introduction
- Coccoid green algae, which reproduce solely through the production of autospores, are among the most complex groups in taxonomy. Based on morphological analysis, these algae were initially classified as belonging to the order Chlorococcales [1]. However, subsequent molecular genetic studies, primarily based on the 18S rRNA gene, have shown that green algae that produce autospores form many independent and unrelated lineages. Most of these lineages belong to the class Trebouxiophyceae or order Sphaeropleales within the Chlorophyceae [2-4].One of the most common types of green algae found in freshwater phytoplankton is Scenedesmaceae, also known as Coelastraceae. These algae consist of flat or curved colonies with oval or spindle-shaped cells. Occasionally, they can form three–dimensional clusters or syncoenobia. Since Oltmann (1904), other authors have significantly revised the scope of this family [1,5]. Currently, the Scenedesmaceae family includes 41 genera: Acutodesmus, Asterarcys, Chodatodesmus, Closteriococcus, Coelastrella, Coelastropsis, Coelastrum, Comasiella, Crucigeniopsis, Danubia, Desmodesmus, Dimorphococcus, Enallax, Flechtneria, Gilbertsmithia, Gloeoactinium, Hariotina, Hofmania, Hylodesmus, Komarekia, Lauterborniella, Neodesmus, Pectinodesmus, Pediludiella, Pseudodidymocystis, Scenedesmus, Sceneoocystis, Schmidledesmus, Schroederiella, Scotiellopsis, Soropediastrum, Staurogenia, Steinedesmus, Tetradesmus, Tetrallantos, Tetranephris, Tetrastrum, Verrucodesmus, Westella, Westellopsis, Yadavaea [6]. Thus, this family is the richest in the order Sphaeropleales, with 367 taxa that have been transferred to new genera due to reclassification of previously known genera such as Scenedesmus, Cohniella, and Crucigenia. Additionally, it includes several genera that have been described de novo, such as Hylodesmus [7], Flechtneria [8], Pediludiella [9].Representatives of the Scenedesmaceae family can be found in various aquatic and terrestrial habitats, including water, soil, stones, sand, lichens, and biocrusts [10-11]. Their isolates have repeatedly demonstrated a high biotechnological potential [12-19] and the possibility of use in monitoring and assessing the quality of freshwater ecosystems [20] and for their bioremediation [21-22].Molecular identification has become an essential and universal tool for algal research [23]. The ITS2 sequence has been widely used as a DNA barcode to confirm the taxonomic identity of freshwater green microalgal strains [24-26]. The aim of this research was to identify and characterize three strains of green algae isolated from phytoplankton samples collected from the Naryn River through a combination of morphological and molecular genetic analyses.
2. Materials and Methods
- Study area and water sampling. Naryn is a river that flows through the territory of the republics of Kyrgyzstan and Uzbekistan in Central Asia. It is formed by the confluence of two other rivers, the Big Naryn and Small Naryn, originating from glaciers in the Central Tien Shan mountains. Naryn has a length of 807 km and a basin area of 59,900 km2, and it plays a significant role in the region's economy, providing water for irrigation and generating hydroelectric power. Water sampling was conducted in 2022 using the Apstein plankton net at two locations (Figure 1):1. Observation point 1 (Nosh) is located on the border with Kyrgyzstan, near the village of Yangiyer (41.160003, 72.145856). The height above sea level is 505 m. Water transparency in the slower parts of the current is 80–100 cm, and in some shallow places, it reaches the bottom. In winter, the water temperature is 0–1.8°C; in summer, the average temperature is 22–23°C.2. Observation point 3 (C01) is located near the bridge–dam in Uchkurgan (41.113258, 72.067386) at an altitude of 490 m above sea level. The water in this area has a transparency of up to 80–90 cm and a temperature of 1–2°C in winter and 22–23°C on average in summer.
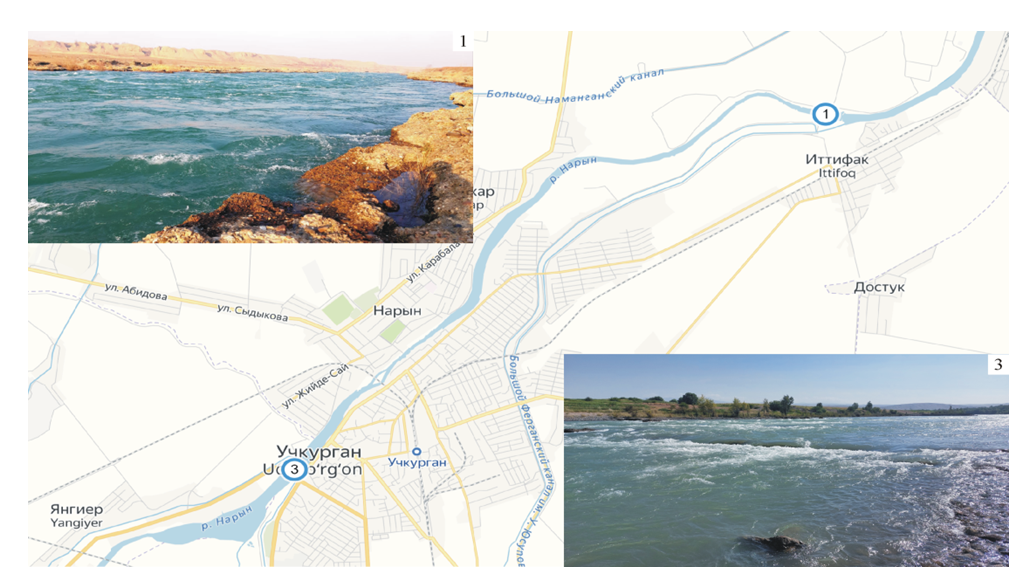 | Figure 1. River Naryn with observation points |
3. Results and Discussions
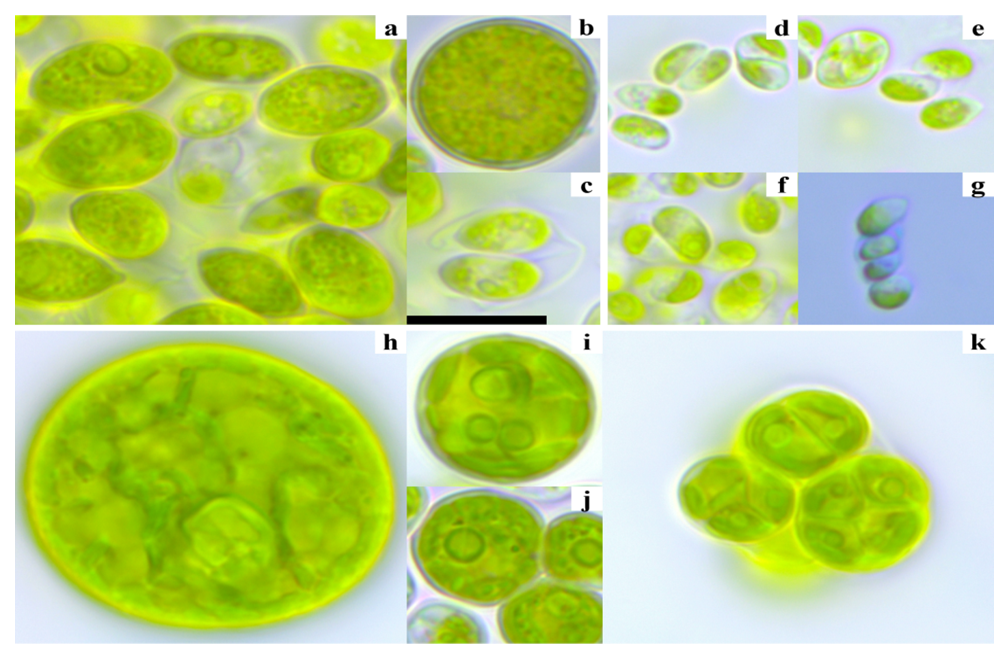
- Three strains of Scenedesmus–like green microalgae were isolated, one from observation point 1 (Nosh) and two from observation point 3 (C01). The morphological descriptions of these strains are provided below.Strain VKM Al–486 from observation point 1 (Figure 2 a–c): The cells are mostly solitary (coenobia were not observed in culture), fusiform, 7.4–11.3 x 4.2–8.1 μm, or very rarely almost spherical–up to 13 μm in diameter. The cell membrane appears smooth under light microscopy (LM), with two (rarely one) polar thickenings. There is a parietal chloroplast with one pyrenoid. Reserve products are numerous grains of starch and drops of oil. They reproduce with 2–4–8 autospores. Remains of the mother cell wall remain in the culture for some time. Sexual reproduction has not been observed. Morphologically, the strain was identified as Tetradesmus obliquus.
4. Conclusions
- Thus, we confirmed for the first time the presence of the species C. tenuitheca, N. danubialis and T. obliquus in the phytoplankton of the Naryn River based on morphological and molecular analyses. The ITS2 as a DNA barcode can be successfully used in the future to monitor the state of freshwater ecosystems. The molecular identification of algae has greatly enhanced our understanding of their taxonomy, phylogeny, and ecology. Accurate taxonomic identification is essential in biology and related fields such as evolution, biogeography, biotechnology and conservation.
ACKNOWLEDGEMENTS
- The author expresses gratitude to the All–Russian Collection of Microorganisms (VKM) staff for their assistance with conducting research and providing consultations.
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML