-
Paper Information
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
International Journal of Virology and Molecular Biology
p-ISSN: 2163-2219 e-ISSN: 2163-2227
2024; 13(4): 54-57
doi:10.5923/j.ijvmb.20241304.02
Received: Jul. 10, 2024; Accepted: Jul. 29, 2024; Published: Aug. 8, 2024

A Comparative Analysis of the Amino Acid Composition of Some Viruses Causing Desease Plum Plants
Sattorov Muzaffar, Fayziyev Vohid
Chair of Biology, Chirchik State Pedagogical University, Tashkent Region, Chirchik, Uzbekistan
Correspondence to: Sattorov Muzaffar, Chair of Biology, Chirchik State Pedagogical University, Tashkent Region, Chirchik, Uzbekistan.
| Email: |  |
Copyright © 2024 The Author(s). Published by Scientific & Academic Publishing.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/

Mutations occurring in phytopathogenic viruses are mainly evident in the gene responsible for their CP amino acid composition and synthesis. Therefore, comparing the CP amino acid sequence of plum pox virus with other isolates is important in solving theoretical and practical problems, such as studying the evolution of viruses. Therefore, in this work, it was aimed to compare the amino acid sequence of CP of PPV with other isolates.
Keywords: PPV, Isolate, Nucleotide, Amino acid, Cover protein, Gene, Phylogenetic tree
Cite this paper: Sattorov Muzaffar, Fayziyev Vohid, A Comparative Analysis of the Amino Acid Composition of Some Viruses Causing Desease Plum Plants, International Journal of Virology and Molecular Biology, Vol. 13 No. 4, 2024, pp. 54-57. doi: 10.5923/j.ijvmb.20241304.02.
Article Outline
1. Introduction
- Today, various strains and isolates of phytopathogenic viruses are being identified all over the world, and their comprehensive study is considered important in the development of countermeasures. In the territory of our republic, the degree of spread of the virus has been determined in several plants, and scientific research is being carried out. In particular, the level of resistance of different varieties and samples of corn to corn mosaic virus, which infects corn plants, was studied [8].A number of features of S, A, M, L, Y and X viruses from the phytopathogenic viruses infecting the potato plant, including the spread of a single potato virus X, biological features such as disease symptoms in indicator-test plants [4,5] and the effect on the amount of chlorophyll "a", "b" and carotenoids in the leaves of Datura stramonium plant was determined [5]. In addition to these, scientific research work is being carried out on the study of potexviruses that infect various agricultural plants even in the climatic conditions of Uzbekistan. In particular, plum pox virus, which infects fruit trees, was first detected in plum plants in the climatic conditions of Uzbekistan [1,6] and its biological characteristics such as disease symptoms in indicator-test plants [6,9] and the virus the effect on the amount of chlorophyll "a", "b" and carotenoids in plant leaves was determined [5]. To date, several isolates of this virus have been studied by a number of authors [1,8], they were found to be different by their molecular-genetic and biological characteristics [3,6], studied populations of isolates of the virus distributed in different regions a number of authors proved this situation based on their experience [8]. Basically, such differences are evident in the process of studying the protein coat (CP) gene of the virus [1,8]. This virus was found to be a new isolate of the virus different from other isolates by nucleotide sequence identification of the ORFs responsible for protein coat (CP) synthesis, and was deposited in International GenBank (NCBI) accession numbers MT038048, MT038050.1 and MT038049.1 based on [6].
2. Materials and Methods
- To date, a number of methods have been developed for determining the amino acid composition of proteins, and they differ from each other in terms of equipment and levels of complexity [10]. Determining the amino acid composition of the viral protein coat was based on sequencing the nucleotide sequence of the gene responsible for the synthesis of this protein coat [7,9], and the sequence of this method is given below. cDNA synthesized by reverse transcription for diagnosis of PPV by polymerase chain reaction (PCR) PCR-mixture (for 1 reaction): 16.5 MilliQ water, 2.5 μl 10x buffer for Taq DNA-polymerase (Evrogen), 0.5 μl of 10 mM dNTP mix, 2 μl of 25 mM MgCl2; 0.5 μl forward and reverse primers (10 pM/μl), 0.5 μl Taq DNA polymerase (PK113L, Eurogen) were added. To prevent evaporation, mineral oil was applied to them. PCR was performed in the Tersik amplifier (DNK-Technology, Russia). PCR sample denaturation (94°C, 3 min), 30 - 35 cycles of amplification, polynucleotide chain recovery 5 - 10 min at 72°C and storage at 4°C. The PCR product required for sequencing was obtained in an analogous manner as above, but the reaction was carried out in 50 μl, the necessary components were calculated and mixed in the same way as above. Alternatively, DNA polymerase Encyclo (Evrogen) was used for accurate transcription. The sitting temperature of the primers and the duration of the elongation process are determined by the characteristics of the primers and the length of the synthesized PCR product [1,8].Analysis of PCR products was determined by electrophoresis on an agarose gel prepared in 1x Tris-acetate buffer (TAE, Thermo Scientific) with ethidine bromide. 3 μl of dye (6x DNA loading dye, Fermentas) was added to 10 μl of the sample and put into the agarose well together with the sample. A mixture of DNA molecules of certain sizes (GeneRuler Plus 100 bp DNA ladder and GeneRuler Plus 1 kb DNA ladder, Fermentas) were used as markers. Electrophoresis was performed using horizon electrophoresis SE-1 (Helikon, Russia) at 80V for 40–60 min. The gel was analyzed at a wavelength of 312 nm on a transilluminator TFP-M/WL (Vilbert Lourmat, France) and photographed using a gel documentation system MultiDoc-It (UVP, UK).Determination of the sequence of the 3'-end of the OCHV genome. The PCR product intended for sequencing was separated on a 1-2% agarose gel. After the end of electrophoresis, the gel was illuminated with a transilluminator TFP-M/WL, the desired area was cut with a scalpel and transferred to a 1.5 ml centrifuge tube. DNA was extracted from the agarose on a spin column using the Cleanup Standard kit (cat. no. BC022, Evrogen) according to the instructions provided by the company. Purified DNA was sequenced using the Sanger method at Evrogen using forward and reverse primers. The sequences of the primers used are listed in the table (Table 1).
|
3. Nucleotide and Amino Acid Sequence Analysis
- Nucleotide and their underlying amino acid sequences were compared using the ClustalW v.2.1 (http://clustalw.ddbj.nig.ac.jp) program or the version included in the BioEdit package [2,9].The obtained comparative information was used to determine sequence divergence and identity and phylogenetic analysis. Phylogenetic analysis was performed in the MEGA6 program using neighbor joining or maximum likelihood and Kimura-2 or Tajima-Nei evolutionary models [1,6,9].Molecular diagnostics of the virus in the MSU "Biochemistry of Plant Viruses" laboratory carried out together with Dr.(Ds), prof. S.N.Chirkov. Therefore, We express our gratitude to Dr.(Ds), prof. S.N.Chirkov.
4. The Obtained Results and Their Analysis
- PPV is one of the strongest phytopathogenic viruses, and it is an RNA capture virus. This virus passes from an infected plant to a healthy plant through mechanical, agricultural machinery [5,9,11]. Together with other viruses, it causes serious damage to agricultural plants. To prevent this damage, the creation of virus-resistant varieties is the most economical and effective way to control OCD. However, it takes a lot of time to breed such varieties, and the mutation of one amino acid can lead to the emergence of new strains.The protein shell is considered one of the important components of the virus, and if the virus particle is visualized in a very simplified way, it can be considered as a shell that surrounds the nucleic acid. As mentioned above, the shell is "capsid" and its sub-elements can be called capsomeres or its morphological subunits. The whole virus particle is called nucleocapsid. In simple viruses such as tobacco mosaic virus, the viral protein coat consists of a single type of polypeptide chain with the same structure. Their amino acid composition is unique to the same protein. In complex viruses (T pair bacteriophages) with dozens of proteins, it is more difficult to determine the composition of all amino acids, because it is characteristic of heterogeneous proteins. In this case, the protein content of each capsid is analyzed [10,11].The amino acid sequence of isolates of OChV MT038048 Uz1 MT038050 Uz2, MT038049 Uz3 isolated in our country was determined based on the ORFs nucleotide sequence and it is presented below (Table 2):
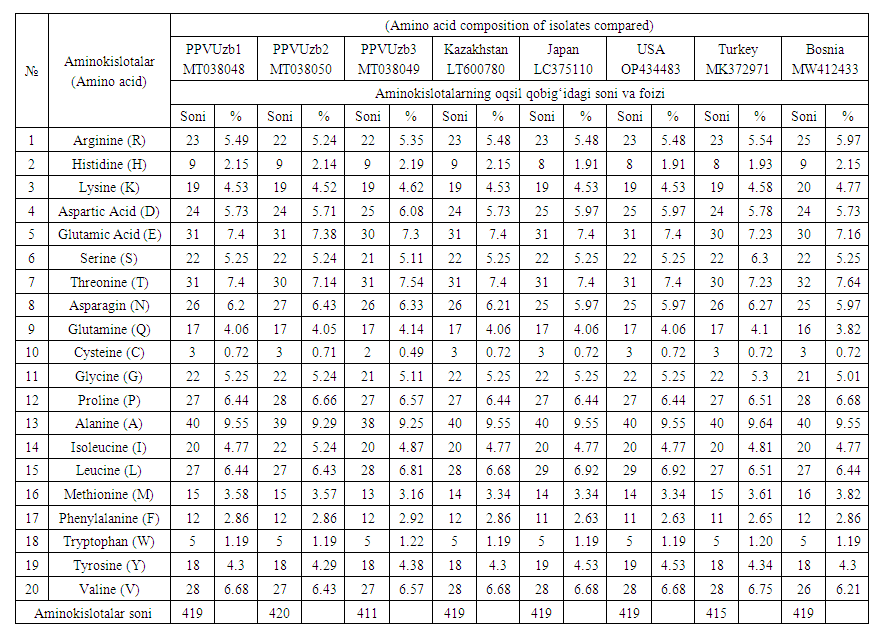 | Table 2. Comparative table of PPV isolate protein shell amino acid composition |
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML