-
Paper Information
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
International Journal of Virology and Molecular Biology
p-ISSN: 2163-2219 e-ISSN: 2163-2227
2023; 12(1): 13-18
doi:10.5923/j.ijvmb.20231201.03
Received: Mar. 27, 2023; Accepted: Apr. 16, 2023; Published: Apr. 27, 2023

Molecular Identification of ToMV Isolates Distributed in Uzbekistan and Study of Some Biological Properties
B. J. Akhmadaliev1, B. A. Abduvaliev1, K. I. Nugmanova1, Z. N. Kadirova2
1Junior Researcher of Institute Genetics and Plant Experimental Biology Academy of Sciences of the Republic of Uzbekistan, Yukori-Yuz, Kibray District, Tashkent Region, Uzbekistan
2Senior Researcher of Institute Genetics and Plant Experimental Biology Academy of Sciences of the Republic of Uzbekistan, Yukori-Yuz, Kibray District, Tashkent Region, Uzbekistan
Correspondence to: B. J. Akhmadaliev, Junior Researcher of Institute Genetics and Plant Experimental Biology Academy of Sciences of the Republic of Uzbekistan, Yukori-Yuz, Kibray District, Tashkent Region, Uzbekistan.
| Email: |  |
Copyright © 2023 The Author(s). Published by Scientific & Academic Publishing.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/

The vegetables consumed by humans, tomato products occupy a special place among vegetables rich in minerals and vitamins. Viral diseases are the main limiting factor in the cultivation of tomatoes and other cultivated plants. Tomato mosaic virus, a member of the genus Tobamovirus of the family Virgaviridae, is the main and most common pathogen of tomato, whose virions are rod-shaped, harboring positive single-stranded RNA. In this study, ToMV was molecularly identified by QT-PCR method, and disease symptoms of ToMV in indicator plants and their appearance periods were determined.
Keywords: Tobamovirus, ToMV, RNA, QT-PCR, Tomato, Mosaic, Necrosis, Nicotiana tabacum, N. glutinosa L., Solanaceae, D. stramonium L., C. annum L., Ch. quinoa L., Ch. amaranticolor L., Ch. album L.
Cite this paper: B. J. Akhmadaliev, B. A. Abduvaliev, K. I. Nugmanova, Z. N. Kadirova, Molecular Identification of ToMV Isolates Distributed in Uzbekistan and Study of Some Biological Properties, International Journal of Virology and Molecular Biology, Vol. 12 No. 1, 2023, pp. 13-18. doi: 10.5923/j.ijvmb.20231201.03.
1. Introduction
- Among the vegetables consumed by humans, tomato products occupy a special place among vegetables rich in minerals and vitamins [1,2]. According to FAO (2009), most of the tomato crops are China, 45 million per year. tons (920.8 thousand ha), USA - 14.14 mln. tons (175.44 thousand ha), Turkey - 10.75 mln. tons (32.46 thousand ha), Ukraine and the south of Russia (from 35-36%), Moldavia (8.4%) and Azerbaijan (5%) from the CIS countries [2].Most of the crops grown as food, fodder, technical and decorative plants around the world are infected with phytopathogenic viruses, which cause significant yield losses and significantly deteriorate the quality of agricultural products. Almost half of the new diseases detected in plants in the last decade are of viral nature [3].Viral diseases are the main limiting factor in the cultivation of tomato and other cultivated plants [4,5]. Virus diseases of the tomato (Lycopersicum esculentum Mill.) plant are one of the main factors that reduce its yield, as a result of infection with viruses, a sharp decrease in yield, as well as a decrease in tomato quality, shelf life, transportability and other properties have been determined [6].Tomato mosaic virus, a member of the genus Tobamovirus of the Virgaviridae family, is the main and most widespread pathogen of tomato, with a rod-shaped, positive single-stranded RNA virion [7]. The ToMV genome, like the genome of other tobamoviruses, consists of four protein-expressing genes, ORF 1 (130 kDa) and ORF 2 (180 kDa) genes are responsible for the synthesis of the replicase, ORF 3 (30 kDa) for the movement protein, and ORF 4 (18 kDa) for the synthesis of the protein coat [8,9,10].ToMV is spread mechanically, through pollen and seeds, or by various virus-carrying vectors (plant aphids, aphids, whiteflies) [11,12]. ToMV causes a number of disease symptoms in plants: mosaic, yellow spotting, curling of leaves, shrinking of leaves, internal necrosis of fruits, etc. [11,13]. Because of ToMV's biological and serological affinity with TMV, Tomato mosaic virus has been considered the "Tomato strain" of Tobacco mosaic virus [13]. Tobamovirus, the so-called "tomato strain" of TMV, with approximately 80% nucleotide sequence similarity to TMV, has been shown to be actually Tomato mosaic virus [14].Based on this, the main goal of this study was to identify ToMV isolates distributed in Uzbekistan based on the replicase gene and to study some of their biological properties.
2. Materials and Methods
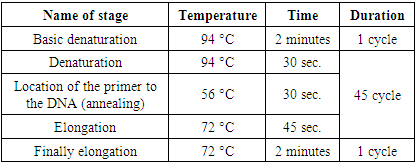
- Virus transmission by mechanical inoculation method. Mechanical transmission of viruses was performed according to generally accepted methods [15]. For this, 100 g of the initially infected plant. and added 0.1 M phosphate buffer (pH 7.2) in a ratio of 1:1 and ground it in a porcelain mortar [16]. The liquid part of the homogenate, which has reached the same mass, is rotated for 15 minutes at 7000-8000 rpm. spun in a centrifuge at 1000 rpm, the supernatant was separated. The isolated supernatant was dusted with carborundum (400 mesh) powder on the leaves of the plant intended for infection, and the virus was introduced into the leaf cells by mechanical microinjury by rubbing the inoculum [17]. After one hour, excess carborundum powder and plant sap were washed off with distilled water. The infected plant was kept in the shade for a day and then brought out into the light. From 2-3 days to 15-20 days, depending on the type of plant, the development of virus-specific disease symptoms was visually monitored from the moment of mechanical transmission of the virus [15,17].Molecular identification of ToMV. The genome of most plant viruses consists of 1-stranded RNA, and their diagnosis by QT-PCR method is carried out in the following steps.Isolation of total RNA. Isolation of total RNA from virus-infected samples was carried out using "PURELINK RNA MINI KIT" kits from "Invitrogen" (ThermoFisher, USA) according to the attached instructions. To do this, samples of tomato plants with ToMV disease symptoms were collected and extracted by placing 5 g in a porcelain mortar and adding liquid nitrogen. 200 mg of the prepared extract was taken, placed in a 2 ml Eppendorf test tube and thoroughly mixed in a vortex (Vortex MX-S, DLAB) after adding 1.5 ml of the lysis buffer (prepared in advance by adding 10 μl of 2-mercaptoethanol to 1 ml of the lysis buffer). Then, it was incubated at room temperature for 3 minutes and spun in a centrifuge for 5 minutes (2600xg rpm). The supernatant was separated and placed in a new test tube. 96% ethanol was added at a ratio of 1:1.5 to the volume of the supernatant and vortexed. It is poured into a filter tube included in the kit, then centrifuged for 15 seconds (12000xg rpm), the precipitate is discarded. 700 μl of washing buffer (Wash buffer I) is added and centrifuged again for 15 seconds (12000xg rpm). The bottom of the column tube was discarded and replaced with a new tube. Then 500 μl of the second wash buffer (Wash buffer II) was added and centrifuged for 15 seconds (12000xg rpm). The filter part of the test tube is removed and placed in a new sterile test tube. 100 μl of RNAse-free sterile water (RNase-Free water) was slowly added from the central part of the test tube and incubated at room temperature for 2 minutes. After the specified time, it was centrifuged for 2 minutes (12000xg rpm) and the filter part of the tube was removed. In this way, total RNA was extracted in the lower tube.Preparation of cDNA by reverse transcription. Invitrogen (ThermoFisher, USA) reagent kit was used for cDNA synthesis based on the isolated RNA matrix. For this, a reaction mixture was prepared by adding ddH2O (2 μl), 5 μl of isolated RNA, reverse primer (GGTGGTGTGCCTCTCTGT) (Integrated DNA Technologies, Belgium) with a concentration of 10 pM/μl and at 70°C for 2 minutes. was incubated. At the end of the incubation time, the mixture was quickly transferred to an ice-cold container. The reaction mixture for obtaining cDNA from ToMV RNA by reverse transcription method includes: reaction buffer-8 μl, dNTP-2 μl, revertase-1 μl, RNA 9 μl. The prepared reaction mixture was placed in the amplifier at 37°C for 60 minutes and 70°C for 10 minutes [14]. Diagnosis of ToMV by QT-PCR method. A reaction mixture (25 μl) for amplification of the synthesized cDNA was prepared as follows: primers selected at 10 pM/μl each based on the NCBI database ToMV-Forward primer-GAGCYTTCACTGAAAGATG (0.5 mkl)ToMV-Reverse primer-GGTGGTGTGCCTCTCTGT (0.5 mkl)Master mix 7.5 μl, ddH2O 12.5 μl, synthesized cDNA 4 μl. PCR amplification was programmed as follows (Table 1) [18]:
|
3. Results and Discussion
- Samples of tomato grown in Andijan, Samarkand and Tashkent regions of Uzbekistan were collected from different varieties and hybrids with mosaic, deformations, virus-like streaks, spots, hardening of the fruit, change of the growth cone of the infected plant, and a number of minor disease symptoms. (Figure 1).
 | Figure 1. Typical signs of viral diseases in tomato plants |
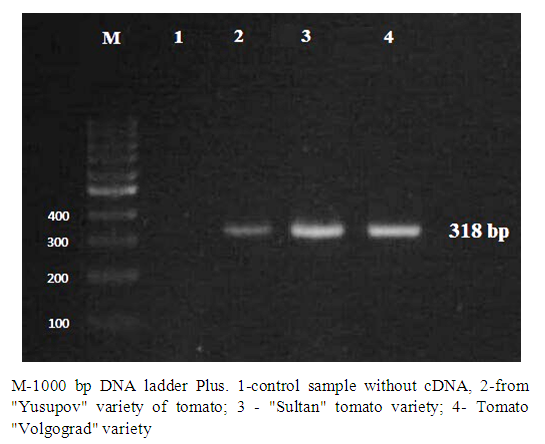 | Figure 2. Diagnosis of ToMV using QT-PCR method |
 | Figure 3. Disease symptoms of ToMV on leaves of Ch. amaranticolor (a), Ch. quinoa (b) and Ch. album (v) plants |
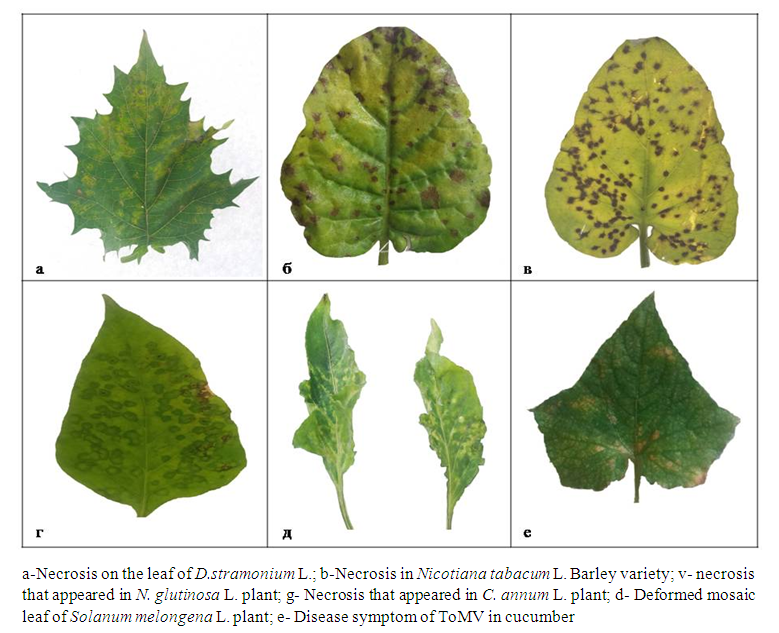 | Figure 4. Disease symptoms of ToMV in indicator plants |
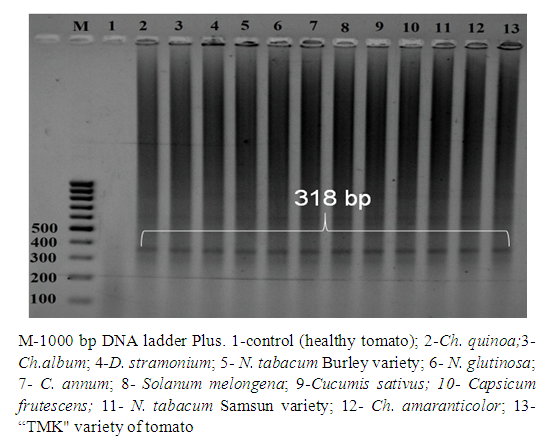 | Figure 5. QT-PCR analysis of ToMV-infected indicator plants |
4. Conclusions
- The presence of ToMV in tomato fields grown in Andijan, Samarkand and Tashkent regions of Uzbekistan was determined using biological and molecular methods. By mechanically infecting ToMV to indicator plants, ToMV infected plants such as D. stramonium L., C.annum L., N.tabacum L., N.glutinosa L. from representatives of the Solanaceae family, and a number of representatives of the Chenopodiaceae family: It was found to cause necroses and chlorotic spots of different sizes in Ch. quinoa L., Ch. amaranticolor L., Ch.album L. species. This study was the first molecular genetic identification of ToMV in Uzbekistan.
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML