-
Paper Information
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
International Journal of Virology and Molecular Biology
p-ISSN: 2163-2219 e-ISSN: 2163-2227
2021; 10(1): 5-11
doi:10.5923/j.ijvmb.20211001.02
Received: Apr. 15, 2021; Accepted: May 17, 2021; Published: May 26, 2021

Comparing the RealStar® SARS-CoV-2 Modified Protocol with Original Protocol to Detect SARS-CoV-2
Asmaa M. Altamimi1, Taghreed A. Alaifan1, Turki M. Alshehri1, Ahmed M. Albarrag1, 2
1Public Health Lab, Public Health Authority, Riyadh, Saudi Arabia
2Department of Pathology, King Saud University, Riyadh, Saudi Arabia
Correspondence to: Asmaa M. Altamimi, Public Health Lab, Public Health Authority, Riyadh, Saudi Arabia.
| Email: |  |
Copyright © 2021 The Author(s). Published by Scientific & Academic Publishing.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/

Background: The use of effective, consistent, and sensitive PCR assays is a foundation to truly identify the virus during this pandemic related to SARS-CoV-2. The SARS-COV-2 laboratory diagnosis depends highly on the RT-PCR technique. This study proposed a new modified protocol (fast) that describes a shorter time profile for the RealStar® SARS-CoV-2 RT-PCR Kit. The study aimed to evaluate the performance of The RealStar® SARS-CoV-2 RT-PCR Kit 1.0 under a shorter time protocol in terms of sensitivity and specificity. Methods: 200 nasopharyngeal SARS-COV-2 samples were subjected to RealStar® SARS-CoV-2 RT-PCR Kit 1.0 test using two different protocols (original VS. fast). We assessed the performance of faster protocol on clinical samples of the RealStar® SARS-CoV-2 RT-PCR Kit for both the E gene and S gene in comparison with the kit's recommended profile. For analysis, the threshold cycle (CT) > 40 or no amplification was considered as negative, while the CT value < 40 with an S-shape amplification curve was considered positive. Results: The results demonstrated the specificity of 100%, sensitivity of 100% using both original and modified protocols. We found that modified protocol helps in getting the results within an hour and 10 minutes instead of two hours, which constitutes a fifty-minute difference. The overall agreement between the two protocols was 100% with negligible differences in the cycle threshold (CT) values for S and E genes. We also found a perfect correlation in the CT values with R2 of 0.98. Conclusions: The modified protocol provides promising results with better sensitivity and specificity in lesser time. Therefore, we suggest a testing strategy that can be done in lesser time and can be used to screen a larger number of individuals.
Keywords: SARS-COV2, Laboratory diagnosis, RT-PCR technique, Fast Protocol
Cite this paper: Asmaa M. Altamimi, Taghreed A. Alaifan, Turki M. Alshehri, Ahmed M. Albarrag, Comparing the RealStar® SARS-CoV-2 Modified Protocol with Original Protocol to Detect SARS-CoV-2, International Journal of Virology and Molecular Biology, Vol. 10 No. 1, 2021, pp. 5-11. doi: 10.5923/j.ijvmb.20211001.02.
Article Outline
1. Introduction
- The coronaviruses belong to the group of large-sized, positively sense, single-stranded RNA with a genome size of 26-32kb, spherical, and non-segmented. The coronaviruses have been grouped into four different genera (γ-gamma, β-beta, δ-Delta, and α-alpha). Out of the four genera in which the coronaviruses have been grouped, only alpha and beta contains CoV-NL63 & CoV-229E and CoV-OC43, CoV-HKU1, respectively. Therefore, it has been proven that the genome of the COVID-19 virus constitutes more than 29,900 nucleotides typically [1]. In many cases, upon mapping and fresh reannotation of such RNA sequences, more than 123,613 of the reader’s assembly will be obtained. They were seen to be similar to those of the SL-CoVZC45, which was an already known strain on the bat, as well as the SARS-CoV [2]. Currently, the total COVID-19 cases, which are recorded globally stand at 105 million, and out of these total cases, 58.5 million have been recovered with the death cases globally stand at 2.29 million people [3].The prevention of the COVID-19 and viral transmission can be inhibited by employing the total lockdown and identification of the infected and asymptomatic individuals at the initial stage to allow early control and a cure. The symptoms were usually confusing with those of the common flu. The asymptomatic carriers will escalate the disease transmission to an uncontrollable manner when they go unidentified and taken to the quarantine. Mass screening of the persons will be necessary for such situations, allowing for selecting those who have contacted the virus. Simultaneously, the detection approaches are typically based on the personal travel history from the areas that are affected; thus, their clinical symptoms are to be identified as well as some of the auxiliary examinations are to be done on such a person [4].Timely Laboratory testing is fundamental in disease diagnosis and control. Clinicians desire precise and rapid results, medical laboratories strive to obtain accurate results within the turnaround time (TAT) and biomedical companies are competing to develop robust assays [5]. During the COVID-19 pandemic, early diagnosis remarkably enables doctors to help patients who are at higher threat for growing more complications from COVID-19. It is critical to have effective testing of disease to distinguish COVID-19 patients from those with different sicknesses. This eliminates pointless isolation of negative people and the spread of the virus by infected patients [6]. The commercially available kits mainly depend on nucleic acid or antibody detection principles [6]. The Real-Time Polymerase Chain Reaction (RT-PCR) approach is considered as the gold standard for SARS-COV2 detection, remains the method of choice, and is approved by World Health Organization (WHO) and Center for disease control (CDC) [6,8]. It depends on targeting a specific sequence of the gene, including nucleocapsid (N), envelope (E), open reading frame (ORF), RNA-dependent RNA polymerase (RdRP), or spike (S) protein [9]. Various approaches aim to reduce the time of testing by amplifying the nucleic acid at a constant temperature to minimize the time of multiple cycles of cooling and heating [10]. We optimized a new protocol condition for The RealStar® SARS-CoV-2 RT-PCR Kit 1.0, and we considered a fast protocol that may work equally well in a short time. This study aims to evaluate the performance of The RealStar® SARS-CoV-2 RT-PCR Kit 1.0 under a shorter time protocol in terms of sensitivity and specificity.
2. Material and Methods
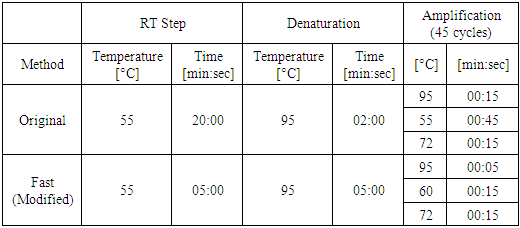
- A total of 200 nasopharyngeal samples previously confirmed were selected (100 positives and 100 negatives for SARS COV-2). 200 µl of each specimen was subjected to extraction using ExiPrep 96 Viral RNA Kit and ExiPrep 96 Lite Automated NA Purification System (Bioneer, Korea). The master mix, reaction setup, and results interpretation were carried out according to the manufactures’ instructions of use. Initially, we aimed to evaluate different cycling protocols on a small number of samples. A pilot study was done on 8 positive SARS-CoV-2 samples with shorter time protocols of RT-PCR steps. Annealing time (10,15,20) seconds, RT time (5,10,15) minutes, and denaturation time (5,10) seconds. Each time we change only one variable of the original protocol and hold the rest and compare the CT values and amplification curves with the same samples on unmodified original protocol. The acceptable values, which gave same results of original protocol in a shorter possible time, combined together as a modified and tested against the original protocol. The acceptable shorter protocol then tested on larger number of samples.The RT-PCR reactions were performed simultaneously on the Light Cycler 480 II instruments (Roche). The cycling protocol for each method is summarized in table 1. For analysis, the threshold cycle (CT) > 40 or no amplification was considered as negative. CT value <40 with an S-shape amplification curve was considered positive.
|
3. Results
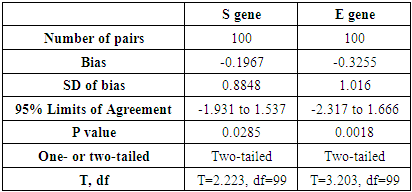
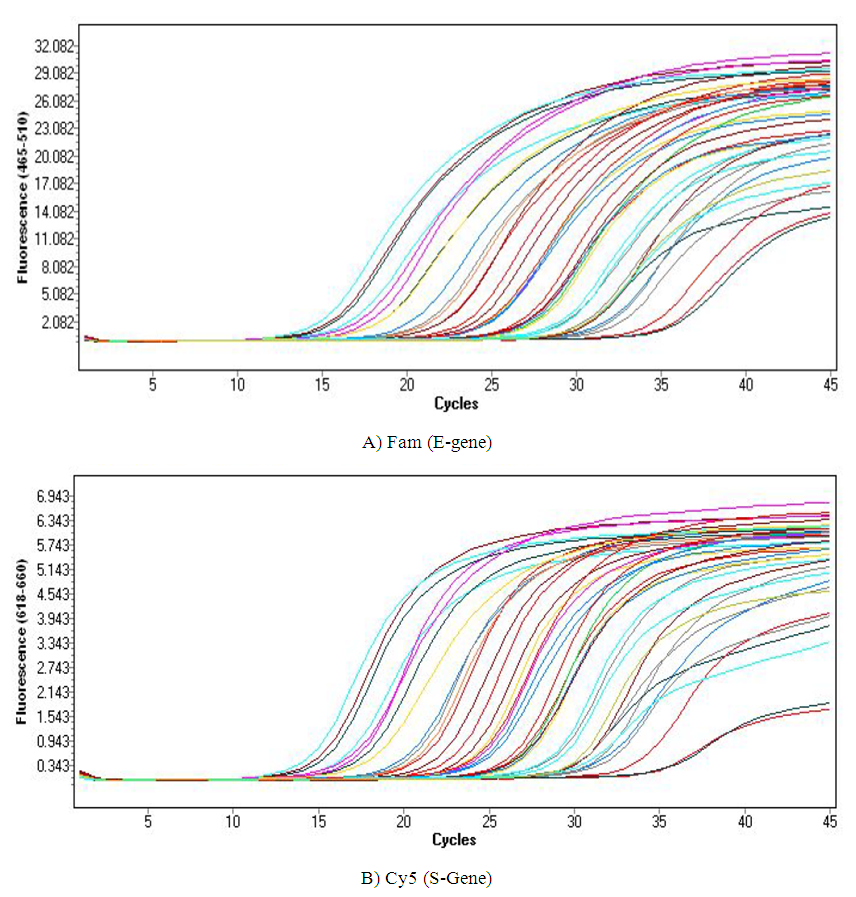
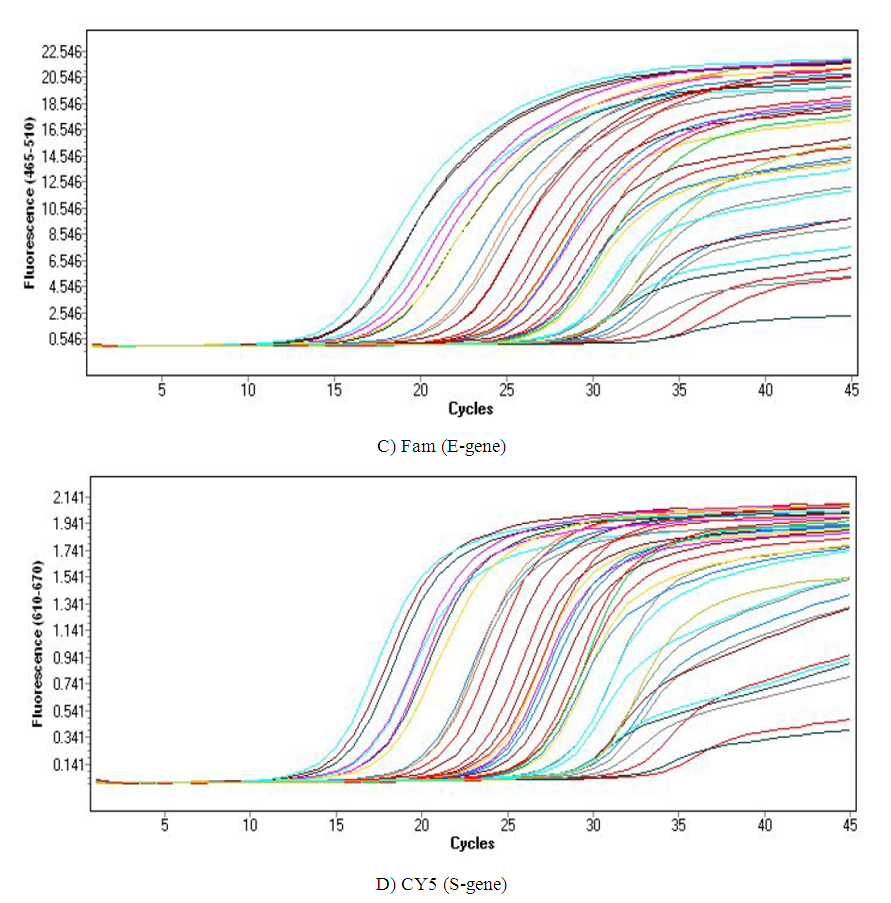
- We used 200 nasopharyngeal samples, 100 each identified positive and negative for both cycling protocols, respectively. While taking the RealStar® kit protocol as the gold standard, our fast protocol demonstrated sensitivity of 100% and specificity of 100% with no false-positive and false-negative results. The fast and original protocols resulted in equally sensitive and specific results with very minor and negligible differences in the absolute values of the CT with an agreement of 100% between the two results. Our data confirmed that both protocols are equal, and the new protocol is conformed with the kit with no compromise on sensitivity and specificity. In addition, we also found that the modified or fast protocol gave similar results as that of the original protocol in about one hour which saved an additional hour to produce similar results.We compared the cycle threshold values for both S and E genes, and the amplification curve shape. Overall, the modified protocol showed better amplification S-shaped curves in comparison to the original protocol see figures (6&7). The wavelength that read the targeted S and E genes were labeled as CY5 and FAM, respectively. The results for S gene showed a negative value for Bias (-0.1967) with a standard deviation of 0.8848 with 95% limits of agreement ranging between -1.93 to 1.537 with significant results (P-value: 0.0285). Likewise, the results for E gene showed a negative value for Bias (-0.3255) with a standard deviation of 1.016 and we are confident that the true value for the estimate falls between 95% limits of agreement ranging between -2.317 to 1.666 with significant results (P-value: 0.0018) as shown in table 2. While assessing the performance of faster protocol on clinical samples of the RealStar® SARS-CoV-2 RT-PCR Kit for S and E genes against the kit's recommended profile, an identical sensitivity and specificity were found for both protocols with 100% agreement. In terms of time duration. It was found that RealStar® SARS-CoV-2 RT-PCR Kit took a shorter time to perform the required number of tests as opposed to the kit's recommended profile. More specifically, the kit's recommended profile took approximately an additional one hour to perform the same number of tests as opposed to RealStar® SARS-CoV-2 RT-PCR Kit.Several graphs were plotted against the cycles of the amplification of the PCR tests (CT value) that helps us to identify the virus and S and E genes for both original and modified protocol. For example, figure 1 reveals the clustering of CT values for the positive samples by the type of protocol.
|
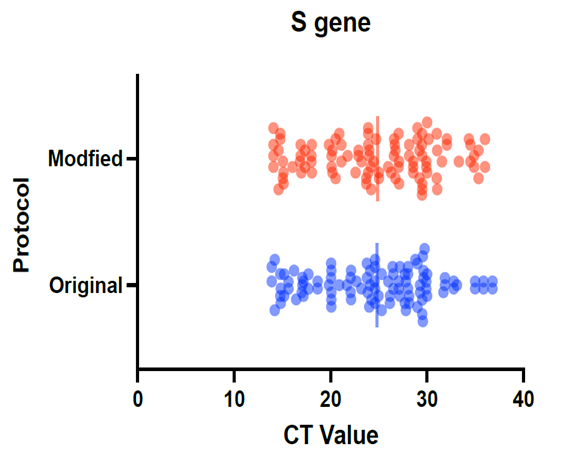 | Figure 1. Clustering of CT values to detect S-gene using modified and the original protocols |
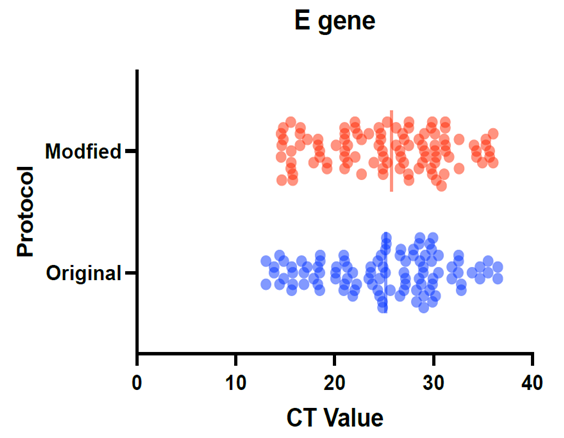 | Figure 2. Pattern for the CT values to detect E-gene using modified and the original protocols |
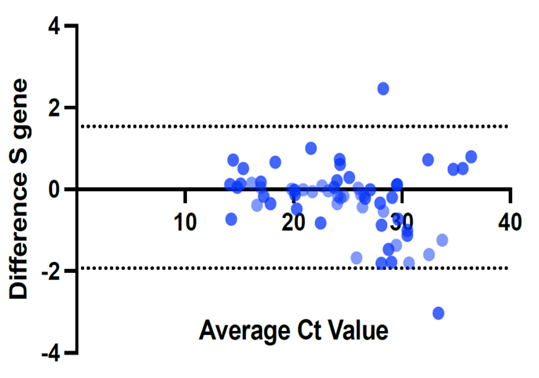 | Figure 3. Difference in the CT values for S-gene using modified and the original protocols |
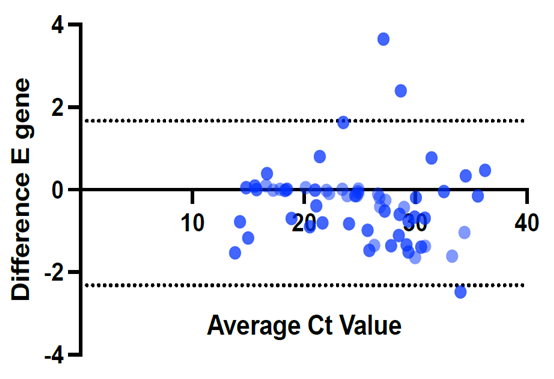 | Figure 4. Difference in the CT values for E-gene using modified and the original protocols |
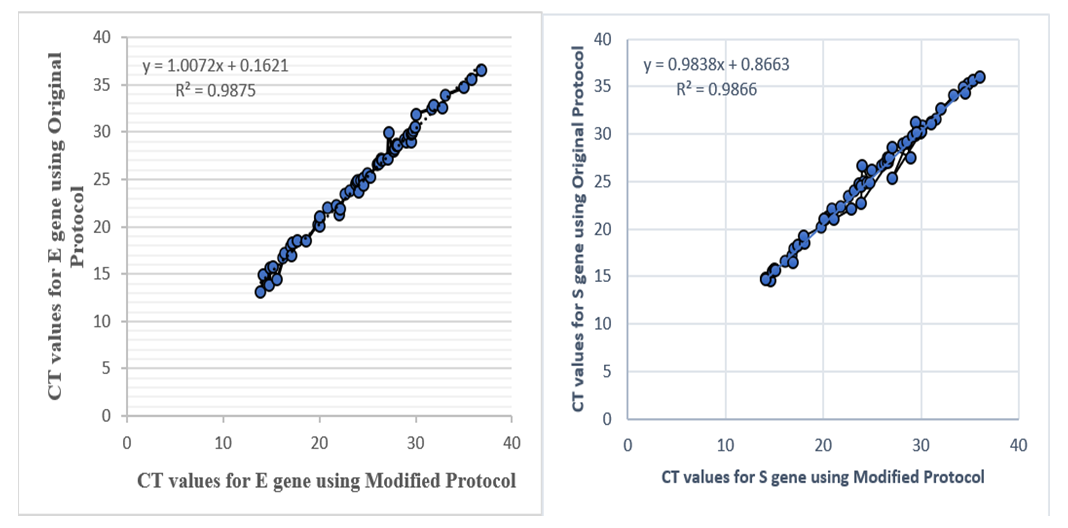 | Figure 5. Correlation in the CT values for S and E-genes using modified and the original protocols |
4. Discussion
- The current pandemic is still a considerable challenge for the healthcare system and policymakers all around the globe. The number of new cases of COVID-19 increases day by day mostly due to asymptomatic carriers [11]. It is also reported that mutation also plays a role in high spread [12]. In European countries, some Asian countries' sudden rise in COVID-19 cases was observed from late September 2020 because of the mutated version of coronavirus. In such a situation, there is an urgent need of the availability of a fast and reliable method for the detection, identification of infected persons. Accessibility and consistency of strong assays offering sensitive identification of diseased people would be considered as the foremost step in isolating the patients with COVID-19, diagnose the same to take the appropriate corrective actions. Furthermore, this will be the first step for the prevention of spread COVID-19 at a greater pace. So far, PCR has been considered as the gold standard test for the detection and confirmation of coronavirus [13]. However, most cases are presented at a late stage, and in some cases, there is a lack of viral antigen [14]. All these points highlight the importance of rapid and highly sensitive PCR assay for the diagnosis of coronavirus. Therefore, we suggested a testing strategy that can be done in lesser time and can be USED to screen a larger number of asymptomatic individuals with high viral loads. The annealing time usually is between 15-30 seconds and for the original protocol it is 45 seconds which is long [15]. We assessed the performance of faster protocol on clinical samples of the RealStar® SARS-CoV-2 RT-PCR Kit for both the E gene and S gene in comparison with the kit's recommended profile. We completed an impartial assessment of the RealStar® SARS-CoV-2 RT-PCR Kit (Altona) to detect SARS-CoV-2. The sensitivity and specificity for both protocols appeared similar with 100% agreement on 200 nasopharyngeal samples. This protocol takes only an hour and 10 minutes, while the original protocol takes two hours. There is about one-hour deference, which helps in performing more tests and releasing more results.
5. Strengths and Limitation
- Our study was the first of its kind that assessed the sensitivity and specificity of a modified protocol in a setting such as Saudi Arabia. This study has contributed to the existing literature to prevent the spread of the virus by timely detecting the disease among other groups of people. Secondly, we conducted an independent and impartial validation of the modified protocol for the detection of the virus and comparing the same with existing conventional method. This in turn gave a pathway to introduce some modified protocols in the laboratories to screen and diagnose the virus. We also carried out the screening and confirmatory tests by detecting S and E genes to make sure that those who were positive for the screening tests were positive on the diagnostic test. However, our study findings need to be interpreted with some caveats such as limited sample size. Main limitation of our study is supply shortage and for future work we aim to assess the efficacy of the modified protocol on larger number of samples. Further study needs to be done to evaluate our finding.
6. Conclusions
- The modified protocol provides promising results with better sensitivity and specificity in lesser time. Therefore, we suggest a testing strategy that can be done in lesser time and can be used to screen a larger number of asymptomatic individuals with high viral loads. The modified protocol offers a vigorous alternative for laboratories working in virology or molecular biology, that can help to detect virus in a smaller time. This will not only save time but also will help to detect the cases quickly that could help to make necessary actions in a timely manner to prevent the spread of the virus.
ACKNOWLEDGEMENTS
- We are grateful to the support of the Public Health Authority, and COVID-19 laboratory Team. Authors would like to thank Ms. Dalia Obeid for her work on figures.
Ethical Approval
- Full ethical approval was obtained by the Saudi Center for Disease Prevention and Control. SCDC-IRB-A023-2021.
Abbreviations

 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML