-
Paper Information
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
International Journal of Virology and Molecular Biology
p-ISSN: 2163-2219 e-ISSN: 2163-2227
2020; 9(2): 40-44
doi:10.5923/j.ijvmb.20200902.03
Received: Oct. 7, 2020; Accepted: Oct. 25, 2020; Published: Nov. 15, 2020

Improving the Methods of Molecular Genetic Analysis in Sports by Studying the Gen ACTN3
Berdiyeva Dilnavoz Toshkan Kizi, Bakieva Gulnoza Habibullayevna, Seidalieva Liliya Tabrisovna, Nishanov Ilyos Burkhonovich, Ziyamukhamedova Sabohat Abdullayevna
Department of Sports Medicine and Biochemistry, Uzbek State University of Physical Culture and Sports, Uzbekistan
Correspondence to: Berdiyeva Dilnavoz Toshkan Kizi, Department of Sports Medicine and Biochemistry, Uzbek State University of Physical Culture and Sports, Uzbekistan.
| Email: |  |
Copyright © 2020 The Author(s). Published by Scientific & Academic Publishing.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/

It is known that the physiological characteristics of a person, such as his structure, strength, speed, endurance and characteristics of the nervous system, are inherited individually and are determined genetically. The main advantage of molecular genetic methods is that a person’s behavioral activity is shown to be hereditary, and early diagnosis reveals a highly informative assessment of the possibility of physical development. A distinctive feature of this diagnosis is the ability to identify hereditary trends, the development of factors of occupational diseases, determining the physical performance of a person and reducing the quality of his life.
Keywords: Sport biochemistry, Gene, ACTN3, Polymorphism, Allele endurance, DNA
Cite this paper: Berdiyeva Dilnavoz Toshkan Kizi, Bakieva Gulnoza Habibullayevna, Seidalieva Liliya Tabrisovna, Nishanov Ilyos Burkhonovich, Ziyamukhamedova Sabohat Abdullayevna, Improving the Methods of Molecular Genetic Analysis in Sports by Studying the Gen ACTN3, International Journal of Virology and Molecular Biology, Vol. 9 No. 2, 2020, pp. 40-44. doi: 10.5923/j.ijvmb.20200902.03.
Article Outline
1. The Relevance of the Research
- Specialists will be able to select young athletes for sports, determining the right direction in sports, the appropriate load, using genetic testing. They can be used as stages in the development of sports genetics in Uzbekistan.
2. Objective of the Study
- To study the importance of the value of gene markers responsible for the development of such qualities as endurance, strength and agility in athletes.
3. Purpose of Research
- Practical introduction of genetic methods in the selection of athletes for sports. Also, molecular genetic study of the endurance marker using genotyping of the ACTN3 gene in R577X polymorphism in athletes.
4. Organization of Research
- This research is based on molecular genetic analysis that will allow an athlete to choose a sport based on his / her genetic, physiological status. This improves performance in sports. Therefore, at the Department of Sports Medicine and Biochemistry of Uzbek State University of Physical Culture and Sports, a theoretical study of molecular genetic analysis was started.
5. Theoretical Analysis of Research
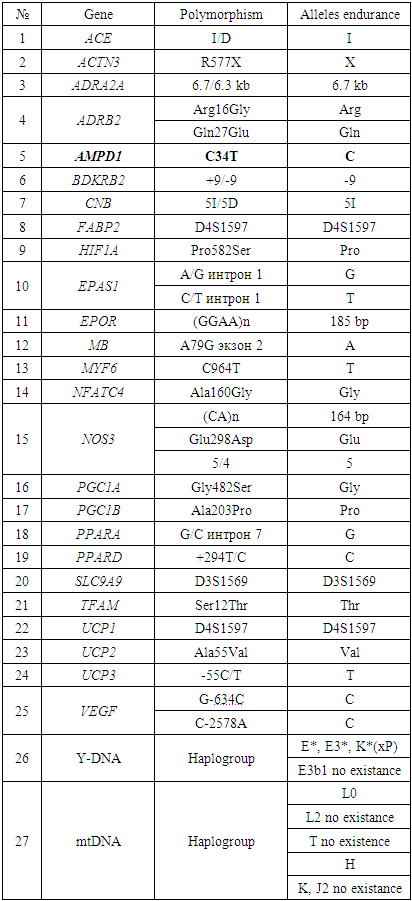
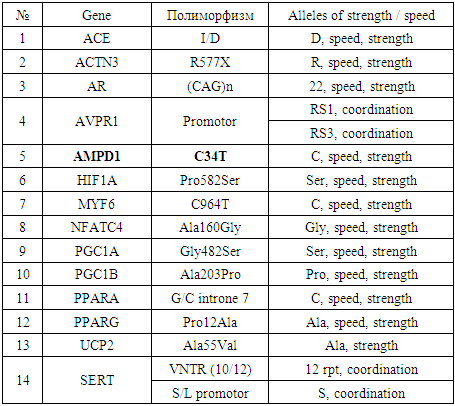
- In sports, genetic methods were first used at the 1968 Olympics in Mexico. Subsequently, in 1976, studies were conducted in Montreal on the differences between Olympic participants and non-athletes. Qualitative genetic markers can easily identify signs of body endurance. Genetic markers can be divided into the following groups:- morphological signs, shape of skeletal muscles, body weight, degree of fat accumulation;- blood groups, erythrocyte antigens; - AVO antigens and leukocytes - HLA;- dermatoglyphic - drawings on the fingers of the pads of the hands and the soles of the feet;- the location and composition of the down bag; - the level of hormones in the blood.Recently, molecular genetic testing has been developed and new opportunities have been discovered for studying sports and molecular genetic problems, as well as optimizing training processes and comprehensive diagnostics (Akhmetov I.I. 2010).Until 2005, data were available on the various genes of 150 people that control physical development (Sharp A.J., 2005). Comparative comparative analysis of the alleles of these genes in different athletes revealed candidate genes and their relationship with different physical characteristics of a person. For example, the following genes identified polymorphic sites of human fitness: polymorphism of the angiotensin converting enzyme (ACE) I / D gene (Wolfarth B 2004, Nazarov I. B 2001), R577X polymorphism of the alpha actinin gene (ACTN3) (Akhmetov II 2010, Sharp AJ 2005, Woods D. 2013), AMP deaminase (AMPD1) polymorphism of the C34T gene (Axmetov I. 2010, Yang N 2013), peroxisome proliferation activator (PPARA), polymorphic site of the alpha receptor and gamma receptor 1-alpha coactivator (PGC1A).Many studies have examined genes for vitamin D receptors (VDR), endothelial synthesis of the gene for nitric oxide (NOS3) and the gene for myostatin (MNN) (Yang N, 2013). At the same time, the alleles associated with the development of endurance (cardiorespirators or muscles), speed-strength qualities, and the development of skeletal muscle hypertrophy were also studied. I allele of the ACE gene and G allele of the PPARA gene are endurance alleles in sports and contribute to high performance. The D and C alleles of these genes are the alleles for "speed" and "strength" (http://sportstati.rf/geneticheskie-markery-sportivnyx-zad/). The R allele of the ACTN3 gene, and the C allele of the PGC1 Age Gly and AMPD1 gene are associated with speed, strength and endurance. Other alleles of these genes have been associated with low physical performance (Mills M.A., 2011). Alpha-2-adenoceptor ADRA2A and genes of mitochondrial DNA haplogroups (mtDNA H) are associated with the ability to limit physical capabilities (Akhmetov I.I., 2010). It should be noted that after the publication of genetic maps of physical activity in 2005, the relationship between gene spectra and the tendency for a specific sport was resolved (Akhmetov I.I. 2010).The list of genes and their alleles associated with endurance and strength (speed) are shown in Tables 1 and 2.
|
|
6. Material and Methods
- In order to study the endurance gene, 50 adolescent athletes and 50 non-athletes were selected. DNA samples were isolated from their venous blood by nucleosorption. When DNA was isolated from venous blood by the colon method, the DNA was not clearly visible on electrophoresis. In the nucleosorption method, however, it was clearly visible.The nucleotide sequence of the ACTN3 gene was searched using the Genbank database. Primers and probes were designed using the online software http://biotools.umassmed.edu/bioapps/primer3 to match the ACTN3 gene. The temperatures of the primers and probes were selected using the programs www.bio.bsu.bu / molbiol / oligocol.html, http://www.basic.northwestern.edu. Current RT (Real-Time RT) reaction on the ASTN3 gene was performed and optimized on DNA samples, and based on the results obtained, the R genotype of the ACTN3 gene was found to be higher and associated in weightlifters than the control group.
7. Research Discussion
- One of the first to study in athletes was chromosome 11 (11q13-q14) localized on the long arm, α-actin-3 (ACTN3, actinin alpha-3). Skeletal muscle α-actinin has two isoforms: α-actinin-2 isoform (ACTN2) and α-actinin-3 isoform (ACTN3), which differ in localization in different areas of muscle fibers. All muscle fibers contain α-actinin-2, while α-actin-3 is localized only in the rapidly twitching fibers of skeletal muscle. A-actin-3 deficiency is a marker characterizing a decrease in the level of human physical activity. The reason for this deficiency of the ACTN3 protein is the replacement of one cytosine nucleotide with thymine at position 577 of the DNA molecule. (point mutation, SNP, R577X). As a result of the mutation, an arginine stop codon is formed, and the synthesis of the α-actinin-3 polypeptide chain is stopped. In the polymorphism of the ACTN3 gene, there are three types of genotypes: normal allele RR-homozygote, heterozygous RX, mutant XX-homozygous alleles. For the X alleles, homozygous individuals lack the α-actin-3 protein in their muscles. In such people, the muscle pathology of the rapidly twitching muscle fibers is compensated by α-actinin-2 (fills the space). In the presence of the normal 577R allele, skeletal muscle contains the α-actinin-3 protein, which confers individual benefits in physical terms such as speed versus strength (Rubio J.C 2005).MA Mills, together with his colleagues, learned to distinguish variants of the three types of the genotype of the ACTN3 gene between athletes and non-athletes. First, homozygote XX has a lower frequency of filling with mutant alleles (7%). Athletes with genotype XX lacking the α-actinin-3 protein in fast twitch muscle fibers have limitations in achieving high results such as speed and strength. In addition, the training of an athlete with genotype XX is very lengthy. As for the owners of the homozygous ACTR3 RR gene and the heterozygous RX, the likelihood that they can achieve high results in sports is high.The next studied gene in athletes is the gene for adenosine monophosphate deaminase 1 (AMFD1, AMPD1), located on the large arm of the 1st chromosome (1p13.1) (Mills M.A 2011). The AMPD1 gene encodes the enzyme adenosine monophosphate deaminase, which is involved in the regulation of energy processes in skeletal muscle. There are three isoforms of AMPD: M (AMPD1 gene, in muscle), L (AMPD2 gene, in liver), E (AMPD3, in erythrocytes). The M AMPD1 isoform is localized to rapidly twitching skeletal muscle. In athletes, a decrease in the activity of the enzyme leads to rapid fatigue or muscle weakness during moderate and intense physical training.The main reason for the lack of the enzyme is the replacement of one nucleotide of cytosine with thymine in the AMPD1 gene (mutation C34T). The AMPD1 gene has three genotypes in the C34T polymorphism: CC is homozygous with the normal allele, CT is heterozygous, and TT is homozygous for the mutant allele.Research by Rubio J.C. showed that 75% of athletes have SS genotypes, 22.6% have a heterozygous CT genotype, and in two studied mutant alleles of a homozygous TT genotype (Woods D 2011). Weightlifting (92%), wrestling (92%) and rowing (70%) prevailed among athletes in the CC genotype. The heterozygous ST genotype was found among athletes in boxers (36%) and speed skaters (36%). In athletes for mutant alleles, the homozygous TT genotype was identified in racers and heavyweight athletes. All of them have low activity of the enzyme adenosine monophosphate deaminase in fast-twitch muscle fibers, which presumably led to high athletic performance. H. Montgomery and co-authors were the first to identify the association of insertion-deletion polymorphism of the ACE gene (I / D) and the growth of athletic performance (Rubio J. C. 2005, Mills M. 2011).The angiotensin gene localized on chromosome 17 catalyzes the transformation of enzymes that convert to angiotensin (ACE), protease, angiotensin-l into angiotensin-II. ACE has been found to be active in several reproductive organs. The enzyme inactivates bradykinin to an inactive metabolite.In the endothelium, Bradokin NO is one of the main stimulants that differentiates the endothelial release factor. Angiotensin II - Bradykinin is a hormone that dilates blood vessels.The ACE gene polymorphism in 16 introns of 287 base pairs is associated with deletion (D) or insertion (I). The ACE gene polymorphism is subdivided into three variants: homozygous I / I, homozygous D / D, and heterozygous I / D genotype. ACE is most active in the homozygote for the D allele. A number of authors have shown that genotype II tends to develop resistance (Rubio J.C 2005). Athletes with the DD genotype tend to react quickly, while the ID genotypes have a high rate of performance. The authors of the above study did not find any differences in genotypes between athletes and non-athletes. But in some sports there is a difference from the control group. For example, in all swimmers the frequency of the I allele is more common than the D allele. On the contrary, in long-distance rowers, the frequency of the I allele is less common than the D allele (Montgomery H. 2009). Hemozygous DD genotypes are common in almost all swimming sports. Middle-distance runners had a higher frequency of I alleles. Marathon runners (in the aerobic type of energy supply) had the highest frequency of the DD genotype. Low concentration of ACE and adaptation of the body to physical activity. Genotype II in relation to the DD genotype of the ACE gene determines 7-8 times higher physical performance (Mills MA 2011). According to many authors, the genotype DD of the ACE gene has a high degree of physical development of speed-strength qualities.In recent years, a family of nuclear receptors, the peroxisome proliferation activator (PPAR), has been studied, which regulates the expression of many genes and is involved in fat and carbohydrate metabolism. These include alpha, gamma and delta receptors, peroxisome proliferation activators (PPARα, PPARg, PPARd). Several studies have shown that one of the PPAR family, PPARγ 1-alpha coactivator (PGC1a), plays a key role in the energy supply of skeletal muscles and myocardium (Montgomery H. 2009). The PPARA, PPARD and PPARG genes are located on different chromosomes. PPARA (Peroxisome Proliferation Activator Alpha Receptor) is localized on the long arms of chromosome 22 of slow-twitch muscle fibers, liver, heart, adipose tissue and energy-generating tissues. Muscle tissue is 7 times more expressive than fat (Rankinen.T. 2010). The main function of PPARα is to provide energy for the processes of lipid and carbohydrate metabolism, control of body weight and inflammatory processes. In physical activity, there is an increase in the use of fatty acids due to PPARα, and PPARα enhances oxidation processes in skeletal muscles (Rogozkin V.A. 2010). One of the key polymorphisms in the PPARα gene is that histidine is converted to cytosine by mutation of the C allele. There are three types of genotypes: GG - abnormal homozygous, GC - heterozygous, SS - mutant homozygous.The PPARG gene (peroxisome proliferation activator gamma receptor) is localized on the shoulder of the chromosome (Zp25). Allele polymorphisms of the Rgo12A1a gene were identified (Nazarov I.B. 2010). As a result of missense mutation, proline is converted to alanine. According to normal alleles, Pro / Pro - genotype, Pro / Ala - heterozygote, according to mutant alleles / Ala / Ala - homozygote are allocated. PPARD (delta receptor for peroxisome proliferation activator) is localized on the short arms of chromosome 6 (6p21.2.1). In this gene, the T / C polymorphism causes a missense mutation and is replaced by cytosine thymine. There are three types of genotypes: TT - homozygous, TS - heterozygous, and CC - homozygous.PGC1A (PPARG gamma receptor proliferation activator) is localized on the large arm of the chromosome (4p15.1). It is expressed from the heart, muscles and adipose tissue. The Gly482Seg mutation most often occurs when glycine is replaced with serine. Gly / Gly - norm, Gly / Ser - heterozygote, Ser / Ser - homozygous for the mutant gene. The frequency of the 482Ser alleles is associated with a decrease in the expression level of the PGC1A gene in 30-40% of the world's population (Shikhova Yu. 2006). In some studies (Rogozkin V.A., 2005, Weyand PG, 2005), the 482Ser allele is associated with obesity, type 2 diabetes mellitus.Most authors compared athletes with a control group for different alleles of different genes. The frequency of the Ala-allele of the PPARG gene and the C-allele of the PPARD gene was increased, the frequency of the Ser-allele of the PGC1 gene was reduced, the indices of the C-allele of the C-PPARA gene did not differ from the control group (Nazarov I.V. 2001, Rogozkin V.A. 2010 , Weyand P.G. 2005, Chen S) 2004). According to information Akhmetov I.I. The G allele activates aerobic potential (activates fatty acid oxidation) and endurance. Allele C. has a more anaerobic potential (increases the processes of glucose breakdown) and leads to the development of speed-strength qualities in athletes (Lucia A. 2005).
8. Research Results
- A comparative analysis of the ACTN3 gene R577X polymorphism genotypes for the distribution frequency of 50 people from 25 weightlifters and 25 football athletes (50 in total) and 50 people in the control group (not regularly engaged in sports) was performed. The R577X genotype of the ACTN3 gene was identified in all (100%) of the 25 weightlifters. No other genotypes were identified in either of the two different groups of athletes. Normal R577X genotype was detected in 37 (74%) of the 50 people in the control group (non-athletes).When the R577X S genotype of the ACTN3 gene is detected, the ascending line of the R graph in the Real-Time (PCR) amplifier rises 5-7 cycles before the X graph. In this case, we can interpret the ACTN3 gene as the X genotype according to the R577X polymorphism (Figure 1).
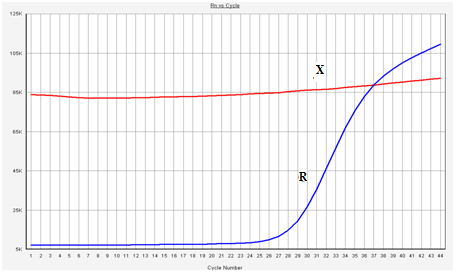 | Figure 1. The result of the current PCR amplification. ACTN3 gene R577X genotype. While the R graph rose from the 24th cycle, the X graph 34 began to rise |
9. Conclusions
- Those involved in sports that require strength and speed should gain muscle mass. Muscle mass is influenced by exercise, nutrition, and hereditary factors.The above mentioned genetic markers are used not only in sports genetics, but also in determining the genetic predisposition to most diseases.In summary, it would be appropriate to include genotyping of the R577X allele of the ACTN3 gene in a comprehensive screening project for those seeking endurance-intensive sports. It is advisable to combine physiological and biochemical analysis, exercise, and psychological conditions to clearly and accurately guide every young person who wants to play sports.
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML