-
Paper Information
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
International Journal of Virology and Molecular Biology
p-ISSN: 2163-2219 e-ISSN: 2163-2227
2020; 9(2): 35-39
doi:10.5923/j.ijvmb.20200902.02
Received: Aug. 27, 2020; Accepted: Oct. 10, 2020; Published: Oct. 26, 2020

Genetic Diversity of Hepatitis B Virus among Blood Donors in Brazzaville, Republic of Congo
Brunel Monic Angounda1, 2, 3, Serge Oscar Mokono1, 4, Luc Magloire Anicet Boumba4, Fabien Roch Niama2, Gabriel Ahombo2, Jean-Rosaire Ibara4, 5, Moulay Mustapha Ennaji3
1National Center of Blood Transfusion, Brazzaville, Congo
2Faculty of Science and Technology, Marien Ngouabi University, Brazzaville, Congo
3Laboratory of Virology, Microbiology, Quality and Biotechnologies/Eco-Toxicology and Biodiversity, Virology Team, Cancerology, Quality and Medical Biotechnology, Faculty of Science and Techniques, Mohammedia University Hassan II of Casablanca, Morocco
4Faculty of Health Sciences, Marien Ngouabi University, Brazzaville, Congo
5Hospital and University Center of Brazzaville, Brazzaville, Congo
Correspondence to: Brunel Monic Angounda, National Center of Blood Transfusion, Brazzaville, Congo.
| Email: |  |
Copyright © 2020 The Author(s). Published by Scientific & Academic Publishing.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/

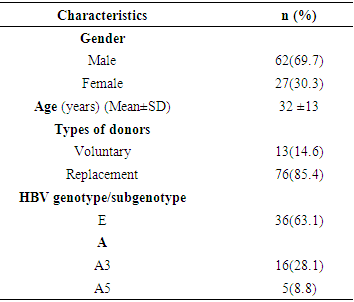
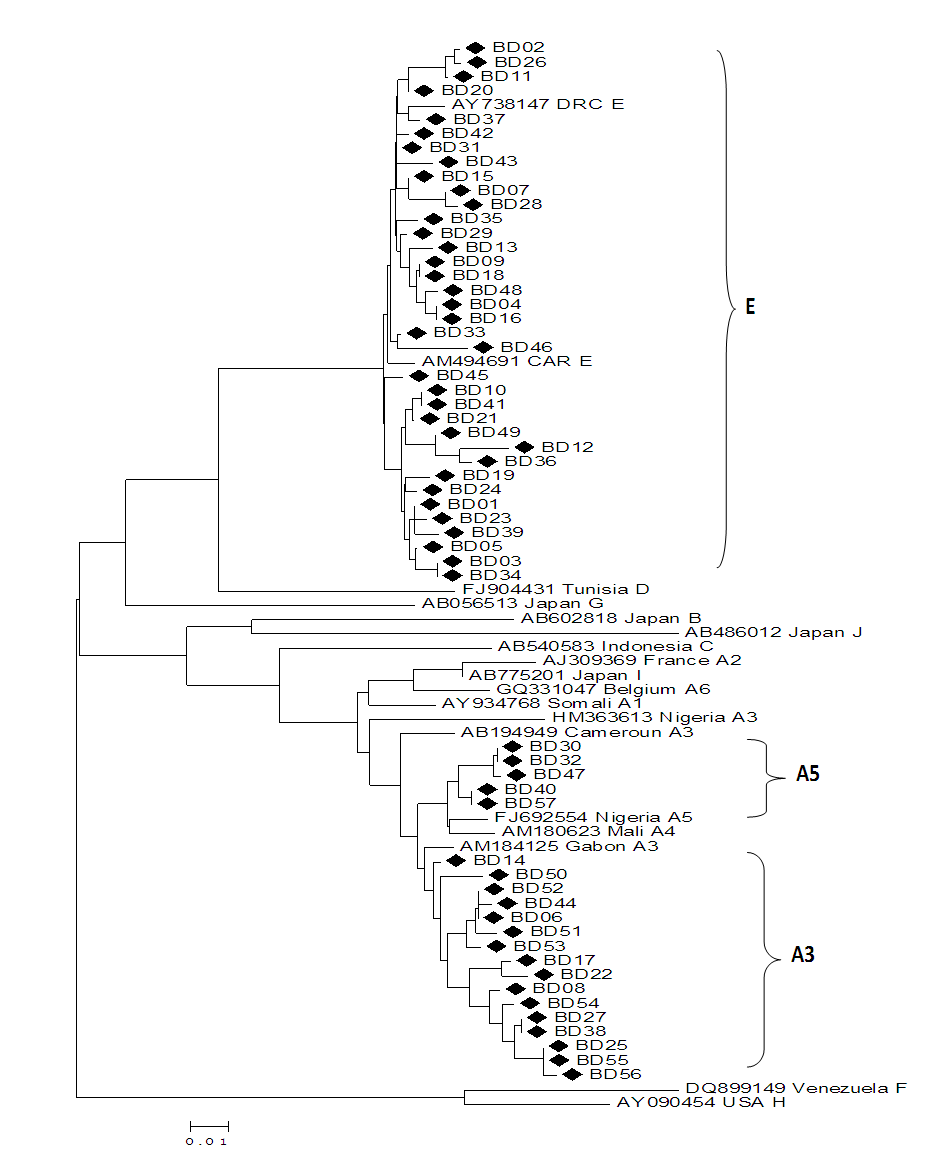
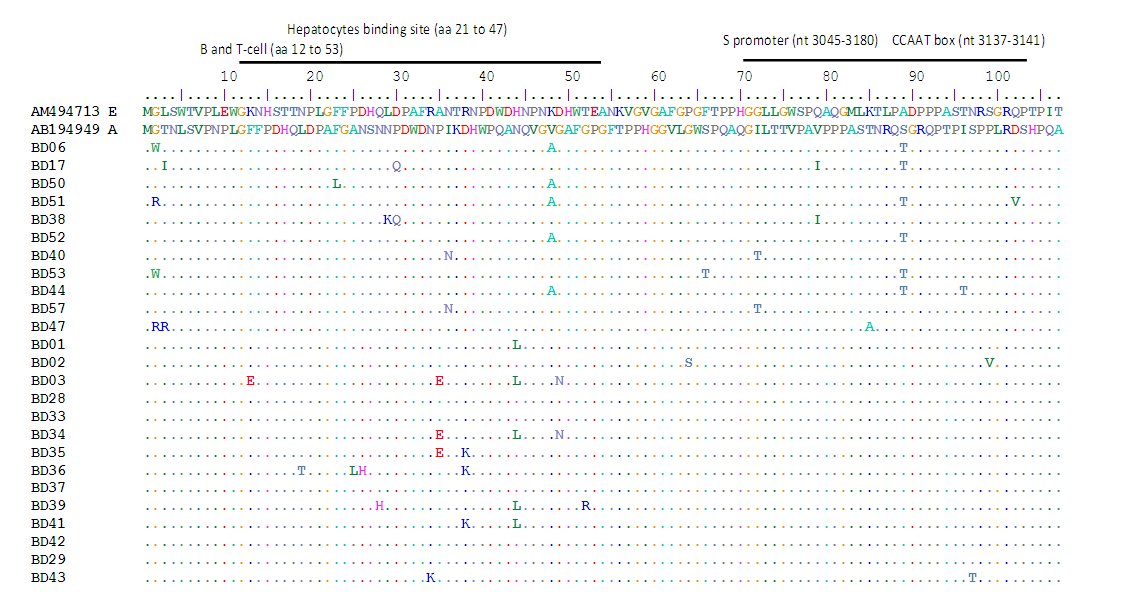
Genetic variations of the Hepatitis B virus (HBV) can affect the disease diagnostic test and influence blood transfusion safety. This study was about the evaluation of the diversity of the HBV strains among blood donors in Brazzaville. A total of 89 hepatitis B surface antigen (HBsAg)-positive samples from blood transfusion center in Brazzaville were enrolled. This was consisted of 69.7% males and 30.3% females with the average age of 32 ±13 years and a range of 18–51 years. HBV genotypes were identified by direct sequencing of pre-S region. Phylogenetic analysis was performed using the BLAST and MEGAv6.0 software. Of 57 amplified samples, HBV/E and HBV/A were detected in 63.16% and 36.84% of the samples. Among genotype A isolates, 76.2% and 23.8% were subgenotypes A3 and A5, respectively. In genotype A the most frequent substitutions were G2W/R, V48A and S89T observed exclusively in 4 (21.5%), 5 (26.3%) and 6 (31.6%), respectively. Multiple mutations A35E, R38K, H44L and D49N were found in 3 (8.3%), 3 (8.3%), 6(13.9) and 2 (5.6%) genotype E infected donors. This study reports the predominance of HBV/E and HBV/A with subgenotype A3 and A5. Phylogenetic analyses revealed that our strains formed a subcluster closely related to Central and West-African sequences previously characterized.
Keywords: HBV, Genotypes, Transfusion, Phylogenetic, Donors
Cite this paper: Brunel Monic Angounda, Serge Oscar Mokono, Luc Magloire Anicet Boumba, Fabien Roch Niama, Gabriel Ahombo, Jean-Rosaire Ibara, Moulay Mustapha Ennaji, Genetic Diversity of Hepatitis B Virus among Blood Donors in Brazzaville, Republic of Congo, International Journal of Virology and Molecular Biology, Vol. 9 No. 2, 2020, pp. 35-39. doi: 10.5923/j.ijvmb.20200902.02.
Article Outline
1. Introduction
- Hepatitis B virus (HBV) infection is a global pandemic disease that affects more than a third of the world population and it is estimated that there are about 350 million carriers of the virus [1]. Of these, approximately 600.000 individuals die annually from complications of the infection [2]. HBV is a partially double-stranded DNA virus of the Hepadnaviridae family and it was classified into ten genotypes designed A to J [3]. Most of the genotypes are further genetically classified into subgenotypes and have distinct geographical distributions [4]. In african countries, HBV genotypes A prevails in Southern, Eastern and Central Africa, genotype D in Northern Africa and genotype E in Western Africa [5]. Subgenotypes have also been identified within genotypes A and D [4,5]. HBV infection is endemic in the republic of Congo, with prevalence of HBV surface antigen (HBsAg) between 7.3 - 15% in various populations [6-8]. Little is known about the molecular diversity of HBV strains in the Republic of Congo. A previous studies conducted on particular areas, have shown that HBV/E and HBV/A are the most prevalent genotypes [6]. Indeed, HBV genotypes may have different pathogenesis and various studies have found an association with disease severity as well as response to antiviral treatment [9]. In addition, the emergence of the variants in the S gene region poses a challenge to the disease diagnosis and risks for blood transfusion safety [10]. Indeed, epidemiology of HBV strains among blood donors help to well-know the levels of HBV carriage and assess the risk of transmission of this infection. However, to date no molecular study on HBV infection in Brazzaville has been reported. Thus, the purpose of this study was to evaluate the genetic diversity of the HBV strains circulating among blood donors in Brazzaville, Republic of Congo.
2. Materials and Methods
2.1. Sample Collection
- The cross-sectional study was conducted at the Brazzaville blood Center from January to June 2014. The plasma samples were collected from the HBV surface antigen (HBsAg) positive candidate of blood donors screened with commercially available kit (Monolisa™ HBsAg ULTRA kit, Bio-Rad, Marnes la Coquette, France). Blood samples were separated into sera and stored at -70°C until use. Informed consent was obtained from all donors for viral testing including the HBV. All the samples used in this study were archived and anonymized leftovers from initial blood testing.
2.2. HBV-DNA Extraction and PCR Amplification
- HBV DNA was phenol/chloroform extracted from 250 μL of serum after the treatment with 0.5 mg/mL of proteinase K for 24 h at 37°C in the presence of 0.2 M NaCl and 0.25% SDS [11]. After precipitation with ethanol, the pellet was dried and resuspended in 30 μL of distilled water. HBV preS1 region was amplified by nested PCR with HBPr1 (nt 2850–2868, 5’GGGTCACCATATTCTTGGG-3’) and HBPr135 (nt 803-822, 5’-CAAGACAAAAGAAAATTGG-3’) as primers for the first PCR and the HBPr2 (nt 2867–2888, 5’-GAACAAGAGCTACAGCATGGG-3’) and HBPr3 (nt 1547–1569, 5’-CCACTGCATGGCCTGAGGATG-3’) for the second PCR as previously [12]. The first-round PCR was performed with Taq DNA polymerase (Promega, Madinson, USA) in a total volume of 25µl, with the following reactions: predenaturation at 95°C for 5 minutes, followed by 35 cycles of 30 seconds of denaturation (95°C), 30 seconds of annealing (50°C), and 30 seconds of extension (72°C), with a final extension at 72°C for 7 minutes. The cycling conditions of the second-round PCR were the same as the first-round PCR but using 2µl of the first-round PCR product as template. PCR cycling was performed on Perkin Elmer 2400 GeneAmpR® PCR thermal Cycler (Scientific Support, Inc, Hayward, CA). The PCR products amplified were subjected to electrophoresis on a 2% agarose gel stained with ethidium bromide-stained and visualized by using an ultraviolet transalluminator.
2.3. Genotype Determination by Phylogenetic Analysis
- Hepatitis B genotypes/subgenotypes were determined by comparing with corresponding sequences of HBV belonging to genotype A–I obtained from GenBank database. Genetic distances were calculated using the Kimura two parameter algorithm and phylogenetic trees were constructed using MEGA version 6 software. HBV sequences were edited, aligned and amino acid substitutions in preS1 region were determined by comparing them against reference HBV genotypes amino acid sequences, using Bioedit version 7.0.4.1.
3. Results
- A total of 89 serum samples from HBsAg positive blood donors were collected. This was consisted of 69.7% males (n=62) and 30.3% females (n=27) with a mean age of 32±13 years and a range of 18–51 years. Among them, HBV DNA was successfully amplified and sequenced in 57/89 (64.05%) samples. The majority were replacement blood donors with 85.4% (Table 1).
|
4. Discussion
- Hepatitis B virus (HBV) infection remains a major global health problem especially in highly endemic areas such as Sub-Saharan Africa, East Asia, and the Amazon basin [5]. In this study, HBV DNA was successfully amplified in 64.05% (57/89) of HBsAg positive blood donors. Indeed, different frequencies of HBV DNA detection were reported in studies performed in deferred donors from Guinea, Sudan and Kenya with 47%, 69% and 65.6% respectively [13-15]. This low HBV DNA amplification rate might be related to poor quality of the purified DNAs or denaturation during storage. However, blood donors have a medical selection before donation and the asymptomatic nature may explain this.We determined circulation of two major genotypes, HBV/E and HBV/A. This result confirms previous reports, which indicated the circulation of HBV/A and the predominance of HBV/E in the Congolese population [6]. Indeed, the HBV genotype E was also predominates in West-Africa on a vast crescent spanning from Senegal to Angola, including Ivory Coast and at least the Western part of The Democratic Republic of Congo [16]. However, it is not clear how far the genotype E crescent extends to the East. This genotype was not found in any other part of the world except for some sporadic cases in African Black in Europe and South America [16,17]. In addition, though slaves have migrated from West Africa to North America, the HBV/E is not found in the USA, suggesting that genotype E emerged from the mid to the late 19th century [18,19]. Additionally, phylogenetic analyses support the splitting of the genotype A into two subgenotypes, namely A3 and A5 when the partial S gene sequences. This finding is similar with study performed in South Africa, in South-Eastern (Tanzania, Malawi) and Eastern Africa (Somalia) where the genotype A is dominating [20,21]. Only the sporadic strains of the genotype A were found in Mali, Burkina-Faso and Nigeria [16,22]. The subgenotype HBV/A1 has been found circulating widely in the continent while HBV/A3, HBV/A4 and HBV/A5 has been reported mainly in West and Central Africa [5,19,22]. Recently, subgenotype HBV/A6 was reported from African-Belgian patients and HBV/A7 from Cameroon [23,24]. This data is important because several authors have reported significant associations between the HBV genotypes and the severity of liver disease, clinical outcomes and the response to antiviral therapies [5,9]. Furthermore, the clinical and virological characteristics may also differ among patients infected with the same genotype [25].The analysis of the Pre-S region shows a genetic variability of our strains. Thus, several substitutions were identified independently of genotypes. Indeed, several lines of evidence indicate a role of the preS deletion mutants in the clinical course of persistent HBV infection [26]. However, the HBV mutants with deletions in the preS1 and the preS2 regions tend to accumulate during a later course of persistent HBV infection, suggesting involvement in viral clearance [27]. Additionally, many studies reported that the HBV genotypes may influence mutations in the pre-S region. In Japanese study, carriers with the pre-S deletion mutants were infected more frequently with the genotype C than with the genotype B and had more advanced disease [28]. On the other hand, mutations in the preS1 region have been proved to a decrease of S promoter transcripts leading to the inhibit HBsAg secretion and may lead to the diagnostic failure [29-31]. This mutation has a significant impact on blood safety, because the detection of core antibody (anti-HBc) and the HBV DNA are not routinely performed. Further studies for evaluating the influences of the HBV genotypes on the emergence of the pre-S deletion mutants are required in a longitudinal observation.
5. Conclusions
- In summary, the present study reports the genetic diversity of the HBV among blood donor in Brazzaville. HBV genotype E was the most predominant, followed by the genotype A with subgenotypes A3 and A5. Phylogenetic analyses revealed that our strains formed a subcluster closely related to the Central and West-African sequences previously characterized. Furthermore, the presence of substitutions in the preS1 region in blood donors, which might influence the diagnosis and should be carefully monitored by further studies including clinical assessment.
ACKNOWLEDGEMENTS
- The authors would like to thank the Congolese Blood Centre staff who participated in the collection and screening of blood donor samples.
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML