-
Paper Information
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
International Journal of Virology and Molecular Biology
p-ISSN: 2163-2219 e-ISSN: 2163-2227
2018; 7(2): 27-32
doi:10.5923/j.ijvmb.20180702.01

Molecular Characterization and Resistance of Hepatitis B Virus to Polymerase Inhibitors in HIV Co-Infected Patients in Abidjan, Côte d’Ivoire, West Africa
Philippe J-L. N’din1, 2, Thomas A. Toni1, Renaud J-J. Dechi1, 2, Léto O. Gogbe1, 3, Jean F. N’guessan1, Emmanuel Brou1, Ahoua Aguia1, Kouadio Kouakou1, Henri Chenal1, Massara Camara-Cisse2
1Virology Laboratory, Abidjan Integrated Bioclinical Research Center (CIRBA), Abidjan, Côte d'Ivoire
2Biochemistry Laboratory, Faculty of Medical Sciences, University of Félix Houphouët-Boigny (UFHB), Abidjan, Côte d’Ivoire
3Laboratory of Pharmacodynamy-Biochemistry UFR Biosciences, University Félix Houphouët-Boigny (UFHB), Abidjan, Côte d’Ivoire
Correspondence to: Thomas A. Toni, Virology Laboratory, Abidjan Integrated Bioclinical Research Center (CIRBA), Abidjan, Côte d'Ivoire.
| Email: |  |
Copyright © 2018 The Author(s). Published by Scientific & Academic Publishing.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/

The co-infection of the human immunodeficiency virus (HIV) with the viral hepatitis B virus (HBV) is one of the major challenges in the management of HIV. This study aimed to evaluate the prevalence of genotypic mutations of HBV resistance in HBV/HIV co-infected patients and to determine the genetic polymorphism of circulating HBV in a population monitored at CIRBA. The primary objective of this study was to assess the antiviral resistance of HBV and to characterize the genetic variability of HBV in HIV/HBV co-infected patients. This is a cross-sectional descriptive and analytical study conducted from January 2014 to September 2016 at CIRBA. Adult patients co-infected with HBV/HIV on ARV but with detectable HBV viral load were included. HBV resistance genotypes were determined by polymerase (pol) gene sequencing performed on the automatic sequencer (ABI 3130 Avant, Applied Biosystems, Courtaboeuf, France). Of 300 HBV/HIV co-infected patients, thirty-nine (39; 13%) were included. The median viral load was 558 IU/mL. The majority of HBV/HIV co-infected patients (n = 36; 92%) were on triple therapy for HIV-1 infection with two tenofovir + Lamuvidine molecules active on HIV-1 and HBV. Analysis of phylogenetic trees constructed using Mega 7 software confirmed that fifteen (n = 15; 100%) were genotype E strains. Resistance mutations (V173L, L180M, M204V, L180M/L, M204M/V) leading to resistance to Lamuvidine, Telbivudine and Entecavir were observed in five sequenced patients (33.33%). This study confirms the circulation of the HBV genotype E in Côte d'Ivoire with an estimated prevalence of genotypic mutation of 33.3%.
Keywords: HBV/HIV Co-infection, Sequencing, Antiretrovirals, Resistance
Cite this paper: Philippe J-L. N’din, Thomas A. Toni, Renaud J-J. Dechi, Léto O. Gogbe, Jean F. N’guessan, Emmanuel Brou, Ahoua Aguia, Kouadio Kouakou, Henri Chenal, Massara Camara-Cisse, Molecular Characterization and Resistance of Hepatitis B Virus to Polymerase Inhibitors in HIV Co-Infected Patients in Abidjan, Côte d’Ivoire, West Africa, International Journal of Virology and Molecular Biology, Vol. 7 No. 2, 2018, pp. 27-32. doi: 10.5923/j.ijvmb.20180702.01.
Article Outline
1. Introduction
- Hepatitis B virus (HBV) infection is a public health problem [32]. Co-infection with HIV is one of the major challenges in its management in resource-limited countries. It is estimated that between 2 and 4 million people worldwide are co-infected with HBV/HIV. In West Africa, this co-infection has a prevalence of 8 to 12% among people living with HIV (PLHIV) [4]. In Côte d'Ivoire, it is 13.4% [5]. To date, this co-infection is an important factor in comorbidity and mortality [9] among people living with HIV.In Côte d'Ivoire, screening for HBV is recommended in the initial assessment with a view to prescribing active molecules on both viruses in the event of co-infection. These prescriptions are made according to the recommendations of the WHO which recommends the use of a triple therapy combining mainly retro-transcriptase and/or protease (PI) inhibitors.Indeed, the nucleoside reverse transcriptase inhibitor (NRTIs) used in this triple therapy in Côte d'Ivoire are also active on HBV polymerase. These are lamivudine (3TC) or emtricitabine (FTC) and Tenofovir Disoproxyl Fumarate (TDF). However, treatment initiation and monitoring should take into account virological tests such as viral load and molecular characterization of antiviral resistance profiles for better management of the infected patient.Several studies were conducted on HBV resistance to polymerase inhibitors in Africa and among these, no resistance to TDF was found [1]. There is very little data available in Côte d'Ivoire on HBV resistance, especially during HIV infection when patients are exposed to these molecules. Thus, in order to contribute to the understanding of HBV genotypic resistance in patients co-infected with HIV/HBV and to improve their therapeutic management, this study aims to determine genetic polymorphism and to evaluate the prevalence of genotypic mutations of resistance in these patients in therapeutic failure in Côte d'Ivoire.
2. Methods
2.1. Study Population
- This is a cross-sectional descriptive and analytical study to assess the prevalence of genotypic HBV resistance mutation in HBV/HIV co-infected patients in Côte d'Ivoire and to determine the genetic polymorphism of HBV circulating in Côte d’Ivoire. All consecutive adult HBV/HIV co-infected patients on ARV therapy with an HBV viral load greater than 6 IU/mL were eligible for the study. Not included in this study were untreated HBV/HIV patients under 18 years of age with a viral load of less than 6 IU/mL. The study was approved by the National Ethics Committee of Life Sciences and Health (CNESVS).
2.2. Determination of HBV Resistance Genotypes
- Viral DNA was extracted after lysis of viral particles and purification on filter columns (QIAamp® Viral RNA Mini Kit (250), Qiagen, Germany) as recommended by the manufacturer. After DNA extraction, the HBV polymerase gene was amplified using the Qiagen Kit (HotStarTaq® DNA Polymerase 1000 Units, Qiagen GmbH Germany) for the first and second PCR according to the manufacturer's recommendations. The primer pairs used were 5'POL m1 186 U 21 (5'- CCCTGC T CGTGGTTACAGGC GG-3') / 3'POLm21174 L 23 (5'-GTTGCGTGCAGCAGCAGCAAA CACTTGG CA-3') for the first PCR and 5'POLm3 251 u 22: (5'- GAC TCGT GGTG GGACTTCTCTCTCA-3') / 3'POL m4 1033L27 (5'-GGCATYAARGCAGATAWCCACA TTG-3') for the second PCR with a concentration of 30µM. The thermal conditions of the first PCR were: 1 cycle at 95°C for 15 min, then (10 sec at 94°C; 30 sec at 55°C; 3 min at 72°C) x 30 cycles followed by 1 cycle at 72°C for 7 min; storage at 8°C. For the second PCR the thermal conditions were: 1 cycle at 95°C for 15min, then (15 sec at 94°C; 30 sec at 55°C; 45 sec at 72°C) x 10 and another cycle (30sec at 94°C; 30 sec at 55°C; (45 sec at 72°C) (+ 5sec /cycle)) x 17 cycles followed by 1 cycle at 72°C for 7 min; storage at 8°C. The final PCR product size was 808 base pair (bp). The amplicons obtained were purified with the QIAquick® PCR Purification Kit (250) (Qiagen, Germany). The sequence reaction was performed with the Bigdye Terminators V3.1 Sequencing Kit (Applied Biosystems, Courtaboeuf, France). The products of the sequence reaction were precipitated and purified by an ethanol purification process according to the manufacturer's recommendations (Applied Biosystems, Courtaboeuf, France). An electrophoretic migration of the purified products was done with the Genetic Analyser sequencer 3130 (Applied Biosystems, Courtaboeuf, France) for sequence determination. The sequences obtained were aligned with the SeqScape 3 software (Applied Biosystems, Courtaboeuf, France) and consensus sequences were aligned with the reference sequences corresponding to the A-H genotypes on the online site http://www.hiv-grade.de/hbv_grade based on the EASL algorithm (European Association for the Study of the Liver) [2].
2.3. Phylogenetic Analyses
- In order to specify the viral subtypes, the consensus sequences obtained were aligned with the reference sequences available in the online site http://www.Hiv-grade.de/hbv_grade. The sequences were aligned with BioEdit v7 software. The phylogenetic trees were made with Mega 7 software [18].
3. Results
3.1. Characteristics of the Patients Included in the Study
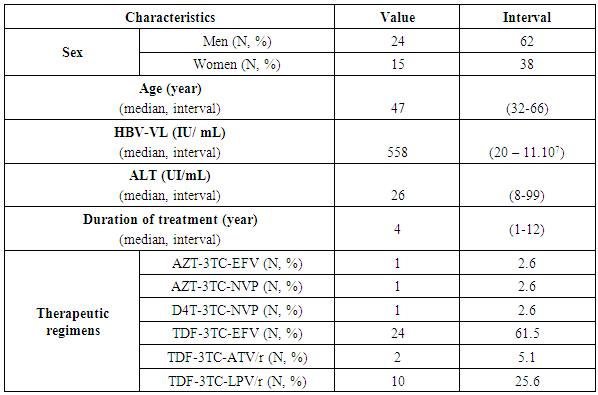
- Thirty-nine patients (N=39; 13%) out of 300 co-infected were included in the study (Figure 1). The median age was 47 years (32 - 66). The median viral load was 558 IU/mL (20 - 11.107) with a value of 26 IU/mL (8 -99) of the enzyme ALanine Amino Transferase (ALAT). The majority of HIV/HBV co-infected individuals (n = 36; 92%) were on triple therapy for HIV-1 infection with two molecules (TDF + 3TC) active on HBV. Three (n = 03; 08%) patients were on triple therapy for HIV-1 infection with one molecule (3TC) active on both viruses. Patients had a median duration of treatment of 4 years (1 - 12) (Table 1).
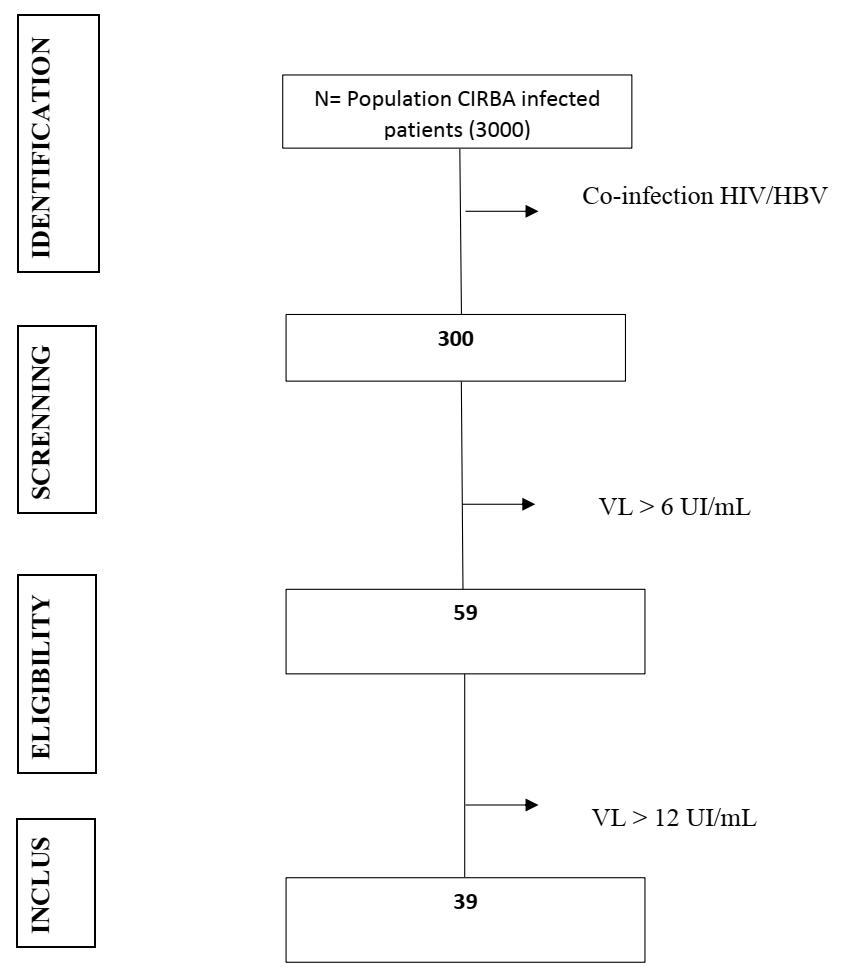 | Figure 1. Identification and selection process for the 39 patients included in the CIRBA study from January 2014 to September 2016. (N = 39/300; 13%) |
|
|
3.2. Distribution of Circulating HBV Subtypes
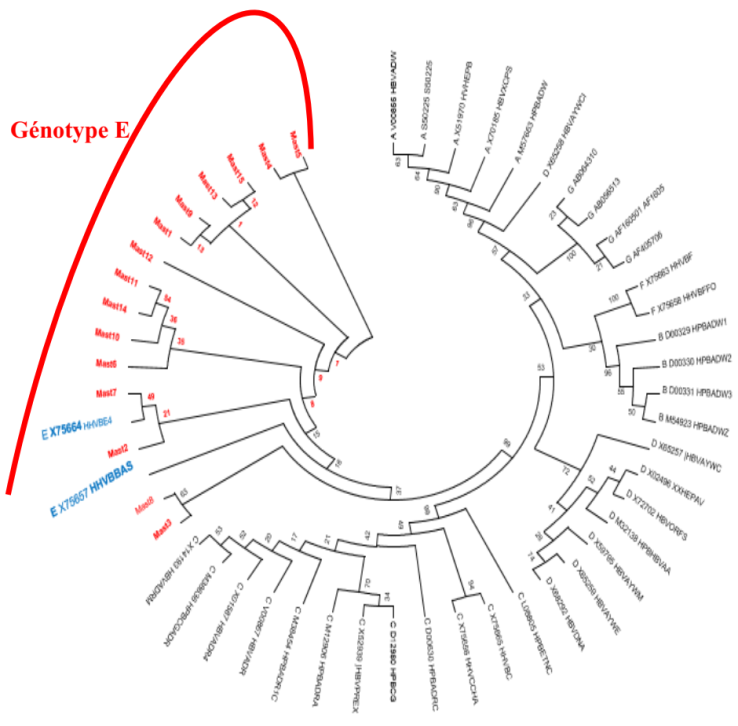
- The pol gene fragments obtained by sequencing were aligned with reference sequences using Bio Edit software. (Figure 2). Phylogenetic trees were made using the Neighborjoining method on the Mega 7 software with HBV reference sequences. Analysis of these trees confirmed that fifteen (n = 15; 100%) strains with positive nested PCR were of genotype E.
3.3. Results of Circulating HBV Resistance Genotypes
3.3.1. Characteristics of Patients with Strains with Resistance to HBV Molecules
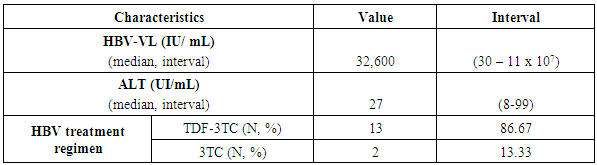
- The number of HIV/HBV co-infected patients with resistance to nucleos(t)idic analogues was (n = 5; 33.33%) on nested PCR positive patients. Three (n = 3; 60%) of patients with resistance to nucleos(t)idic analogues used the combination TDF/3TC and only two (n = 2; 40%) used 3TC as molecules with activity on both viruses. The median duration of treatment was 9 years (1-12) with a median viral load of 2.76 x 107 IU/mL (Table 3).
|
3.3.2. Resistance Mutation of HBV
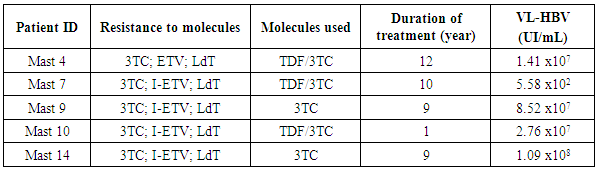
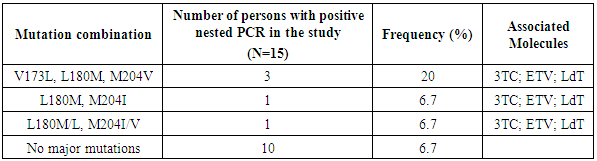
- Resistance mutations were observed in five sequenced patients. On the reverse trancriptase (RT) domain of the HBV polymerase gene, V173L, L180M, M204V resistance mutations were observed. The combination of these resistance mutations was specific for 3TC, ETV and LdT in three (n = 3; 20%) of sequenced patients. On the same gene, mutation combinations such as (L180M, M204I) and (L180M/L, M204I/V) were observed in two patients (n = 2; 13.4%). Thus, the prevalence of mutation resistance in 3TC was 33,3% (Table 4).
|
4. Discussion
- In this study, we showed that the prevalence of HIV/HBV co-infection remains high (10%) and confirms data from previous studies conducted in Côte d'Ivoire [5].The prevalence of co-infection remains high among men (n = 15; 41%) with a sex ratio of 1.6 men to 1 woman. These results are superimposable with those reported by Doumbia et al., [12]; Mukhlid et al., [22]; Traoré et al., [26] and confirm the high prevalence of HBV/HIV co-infection among men.Regarding antiviral coverage, only twenty-four 24 (24; 61%) of patients had active 3TC and TDF therapy on HBV. The adoption and implementation of the WHO 2016 [28] and PNLS 2015 [24] recommendations for the management of HIV/HBV co-infected patients, which stipulate that the combination of 3TC and TDF in triple therapy will allow for greater openness as described in a study in Mali Traoré et al. 2014 [26].For HBV viral replication in virologic failure patients, the median was 558 IU/mL [20 IU/mL; 1,1.108 IU/mL]. This replication would be the consequence of insufficient drug pressure due to poor ARV intake [17] or selection of resistant viral variants.Phylogenic analysis of the sequences obtained showed that 100% of the strains were subtype E. These results confirm the predominance of this genotype in Côte d'Ivoire. [12, 15, 16] But this study does not exclude other HBV genotypes in West Africa such as genotype A that have been found in pregnant women in Mauritania [20] and Senegal [27].As for the resistance genotypes, the comparison of our sequences to the reference sequences confirms the presence of mutation at M204V (n=3; 23%), L180V (n=4; 30%), L173V (n=3; 23%), M204I (n=1; 8%), L180M/L (n=1; 8%) and M204I/V (n=1; 8%) which are mutations of resistance to 3TC, ETV, and LdT [29].On the HBV polymerase gene, the most common mutations were L180M, M204V and L173V. The L180M mutation was observed in 4 patients, while the M204V and L173V mutations were found in 3 patients each.With respect to these resistance mutations, the M204V mutation was identified in a study on the clinical consequences of HBV antiviral resistance [14]. They considered that this mutation was one of the main mutations that induced resistance to 3TC.In another recent study, M204V and L180V mutations were described as a combination of HBV polymerase resistance mutations to both 3TC and LdT molecules [10].A study on the integration of sequence analysis tools and application to modelling confirmed the presence of M204V, L180V, L173V mutations [13]. These mutations are associated with resistance to 3TC, LdT and ETV.These data show an estimated 20% increase in the prevalence of HBV resistance in 2008 [25]. This increase could be related to greater access to antiviral treatments in the HIV program and HBV infection monitoring.In HBV monotherapy patients (n=2; 40%) with 3TC, we observed a virologic failure characterized by a high HBV viral load. A study conducted in Tunisia confirmed this prevalence of 3TC resistance as monotherapy at 42% after 2 years of treatment [6]. In patients on HBV dual therapy (n = 3; 60%) we observed lamivudine resistance but no resistance to TDF despite high viral loads. Failure with lamivudine could be due to taking these molecules over several years as demonstrated in several previous studies [8, 11].In this study, no resistance to TDF was described despite the median treatment duration estimated at 9 years. This was confirmed by Bihl et al., 2015 [9] in a study conducted in Switzerland including 143 HBV/HIV co-infected patients treated with TDF.Studies have shown that the most common mutations resulting in 3TC resistance occur in particular in the highly preserved "YMDD" (tyrosine methionine aspartate) pattern of the catalytic domain (C domain) of the viral DNA polymerase [31].Methionine substitutions for codon 204 to isoleucine (rtM204, YIDD variant), valine (rtM204V, YVDD variant) or serine (rtM204, YSDD variant) are the predominant mutations causing 3TC resistance [21, 31]. In addition to the selection of the "YMDD" pattern, the frequent association of rtL180M mutations and rtV173L was observed in the B domain of HBV polymerase [30]. In the 5 patients with an antiviral resistance profile, we found that in all (n=5, 100%) there was at least one mutation from the YMDD pattern. This high rate of YMDD mutations has been demonstrated in several studies. In Li et al. 2006 [19], it was found that in 142 HBV positive patients treated with 3TC for 2 to 4 years, 56.3% of patients had YMDD mutations; in the same context another multicentre study conducted in China in 1042 HBV positive patients by Tan et al. 2012 [24] showed that 23.3% (243/1042) of these patients had resistance mutations from the YMDD motif.
5. Conclusions and Recommendations
- This study on the molecular characterization and resistance of the hepatitis B virus to HBV polymerase inhibitors characterized the main strain of HBV circulating in a population of PLHIV in Côte d'Ivoire. It also made it possible to evaluate antiretroviral resistance in patients co-infected with HBV/HIV under triple therapy by implementing a genotyping technique by sequencing the polymerase gene.Phylogenetic analysis of the sequences obtained confirmed the circulation and predominance of the E genotype in Côte d'Ivoire. Significant resistance mutations have been observed in the HBV polymerase gene in some virologically failed patients. This allowed us to estimate the prevalence of genotypic mutation in our study at 33.3%. These mutations could be involved in the resistance of HBV to certain antiretroviral molecules belonging to the class of HBV polymerase inhibitors such as 3TC, LdT and ETV. TDF remains an active molecule and should be the first-line molecule for the treatment of HBV.HBV genotyping by sequencing is available and will allow epidemiological surveillance.
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML