-
Paper Information
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
International Journal of Virology and Molecular Biology
p-ISSN: 2163-2219 e-ISSN: 2163-2227
2017; 6(1): 1-8
doi:10.5923/j.ijvmb.20170601.01

Occurrence of Bacillus thuringensis Bacteriophages in the Egyptian Arid Soil
Amel S. M. Abo-Senna
Botany and Microbiology Department, Faculty of Science (for girls), Al-Azhar University, Cairo, Egypt
Correspondence to: Amel S. M. Abo-Senna, Botany and Microbiology Department, Faculty of Science (for girls), Al-Azhar University, Cairo, Egypt.
| Email: |  |
Copyright © 2017 Scientific & Academic Publishing. All Rights Reserved.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/

In this study, two phages specific for Bacillus thuringiensis were isolated from arid soils. The phages were designated as Bt1 and Bt2. They were propagated by the liquid enrichment method and purified by the polyethylene glycol and dextran sulfate followed by high speed centrifugation, the morphology of purified particles was determined using transmission electron microscopy. Electron micrograph indicated that Bt1 phage has isometric particle, its head diameter is about 140 x165 nm and contractile tail type, about 117 nm length, while the particle of phage Bt2 has long contractile tail with 206 nm length and its head is isometric with diameter of about 140 x 124 nm, phages appeared to be a member of the Myoviridae family based on their structure and particle morphology. The isolated phages have narrow host range, which they able to infect Bacillus thuringiensis, Bacillus subtilis and Bacillus lechiniformis, Bt1 and Bt2 viral genomes were digested by EcoRI. The genome size was estimated to be ∼4.06 kbp for Bt1 phage and ∼6.43kbp for Bt2 phage. To our knowledge, this is the first report on the isolation of a bacteriophage specific for Bacillus thuringiensis from arid soil in Egypt.
Keywords: Bacillus thuringiensis, Phage morphology, Purification, Restriction enzyme
Cite this paper: Amel S. M. Abo-Senna, Occurrence of Bacillus thuringensis Bacteriophages in the Egyptian Arid Soil, International Journal of Virology and Molecular Biology, Vol. 6 No. 1, 2017, pp. 1-8. doi: 10.5923/j.ijvmb.20170601.01.
Article Outline
1. Introduction
- Desert biomes cover 33% of global land masses, and yet the diversity and roles of viruses in these dominant ecosystems remain poorly understood. There is evidence that hot hyper arid desert soils are characterized by high levels of bacterial lysogens and low extracellular virus counts [1]. Research on phages ecosystem ecology in hyper arid desert soils has demonstrated that desert soil viruses are numerous and diverse and carry novel genes whose functions are yet to be determined. Bacteriophages are the most abundant biological entities on the planet; their ability to infect and kill their bacterial hosts makes them key factors in both the evolution of bacteria and the maintenance of ecological balance [2-4]. Research on the virus ecology of soil environments has progressed more slowly and has received proportionally less attention [5-7].Desert soil microbial ecology research has primarily focused on bacterial communities, which have been shown to be largely responsible for primary production and provision of key ecosystem services [8-11].Bacillus thuringiensis (Bt) is gram-positive, naturally occurring soil bacterium that produces proteinaceous endotoxins, known as crystal inclusion (cry) protein, which are highly toxic to insects and pests [12, 13]. More than 60 cry genes, with toxicity spectra extending over several, the natural occurrence of B. thuringiensis in soil environments was reported from Egypt [14] and Jordan [15]. However, sensitivity of Bt to lysis by bacteriophages is the most one of complicated to such industry. Lysis can either be caused by exogenous phage infection or be due to strain lysogeny. About 83% of Bt subspecies contain lysogenic phage [16].Bacteriophages are a diverse group of viruses that infect bacteria and are found in a variety of environments, which include soil, drinking water, food, and so on [17]. It is estimated that the biosphere consists of approximately 1031 bacteriophages and archael viruses. Bacteriophages are known to proliferate wherever their bacterial hosts exist [18]. Bacteriophages are the most numerous form of life on earth; ten times more numerous than bacteria [19, 20]. They can be found in all environments where bacteria grow: in the Sahara, hot springs, the North Sea, and polar inland waters [21-23]. Phages are detected in ground and surface water, soil, food (e.g. sauerkraut, wine), sewage, and sludge [24-26]. They have also been isolated from humans and animals, for example from feces, urine, saliva, spit, rumen, and serum [27-29].Bacteria and their viruses (called bacteriophages, or phages), have been found in virtually every ecological niche on Earth. Arid regions, including their most extreme form called deserts, represent the single largest ecosystem type on the Earth's terrestrial surface [30]. This research aims to investigate the presence of bacteriophages specific for Bacillus genus specially B. thuringiensis in the arid soil because the researches in this field are very few and need the more to add much information about the occurrence of bacteriophages.
2. Materials and Methods
2.1. Soil Samples
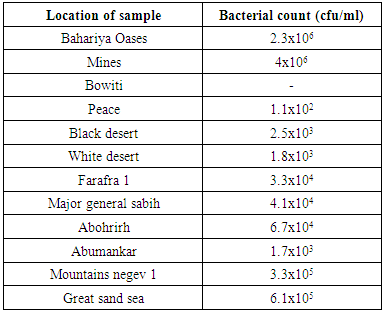
- Twelve soil samples were collected from different locations of the Egyptian desert as arid soil. Four samples from Giza governerate (Bahariya Oases, Mines, Bowiti, Peace) and eight samples from black desert, white desert, Farafra 1, major general sabih, Abohrirh, Abumankar, Mountains negev 1 and Great sand sea were investigated, and Enumerated for total count of bacteria.
2.2. Source of Bacteria
- Different identified species of Bacillus subtilis, B. thuringiensis, B. polymyxa, B. megaterium, B. amyloliquefaciens, B. lechiniformis and B. circulans were kindly obtained from virology lab., Agricultural microbiology Dept., Faculty of Agriculture, Ain Shams University, Cairo, Egypt. All bacterial cultures were sub-cultured and maintained on nutrient broth and nutrient agar medium.
2.3. Enumeration of total Count of Bacteria
- Tryptic soy agar medium was used to enumerate the total count of bacteria in all collected samples by plate count technique. The plates were incubated for 48h at 30°C.
2.4. Preparation of the Crude Phage Lysates
- The specific phages for Bacillus sp., were detected in different arid soil samples using the liquid enrichment technique, method of Duff and Wyss [31] modified by Othman [32]. Five grams of soil were added to 50 ml of nutrient broth medium was inoculated with a mixture of different bacillus strains were used (Bacillus subtilis, Bacillus thuringiensis, Bacillus polymyxa, Bacillus megaterium, Bacillus amyloliquefaciens, Bacillus lechiniformis and Bacillus circulans) in 250 ml Erlenmeyer flask with traces of CaCO3. Then, flasks were incubated at 30ºC for 72 hr with shaking (250 rpm/min). After incubation the cultures were centrifuged at 6000 rpm for 15 min and the supernatant was collected in a clean flask. Chloroform was added with rate of 1:10, followed by vigorously shaking for 5 min. The crude lysate of the phages was obtained and assayed qualitatively and quantitatively.
2.5. Qualitative and Quantitative Assaying of Bacillus Phages
- Bacillus phages were qualitatively assayed by the spot test technique according to the method of Othman [32]. Quantitative assaying of the phages was carried out by over layer agar technique according to Adams [33] and as reported by Othman et al. [34]. For the quantitative assaying, phage dilutions were made in nutrient broth and plaque assay technique was done.
2.6. Preparation of the Phage Isolates
- The different isolates of phages specific for Bacillus thuringiensis were performed by a single plaque isolation method based on the plaque morphology. Single characterized plaques of Bacillus thuringiensis phages were characterized and picked up, then transferred to flasks containing 10 ml of liquid host culture (108 cfu/ml), followed by incubation at 30°C for 48-72 hr with shaking at 150 rpm using a shaker incubator. The single plaque isolation was repeated three times until a high degree of plaque purity was achieved [35].
2.7. Preparation of High Titer Phage Stock Lysates
- A large amount of the phage stocks were obtained by propagation the phage by the liquid propagation method used by Othman et al. [34].
2.8. Purification and Concentration of Bacteriophages
- Dextran sulfate-polyethylene glycol two phase liquid system was used [34]. To purify and concentrate the bacteriophages of Bacillus thuringiensis phage lysates, the weights 222.3, 0.48, 15.8 and 4.2 grams of phage lysate, dextran sulfate 500, polyethylene glycol (PEG 6000) and NaCl, respectively were mixed in a separating funnel to give a mixture containing ratio 6.5%, 0.2% and 1.7% (W/W) of PEG 6000, dextran sulfate and NaCl, respectively. After mixing the funnel was allowed to stand at 4°C overnight. A heavily turbid bottom layer was slowly collected into a clean tube and centrifuged at 2000 rpm for 10 min. The clear top and bottom phases were removed by pipette and the remaining interface "cake" was suspended in 2.5 ml of a 1% (W/W) dextran sulfate solution. Then, 0.15 ml of a 3M KCl was added to each milliliter of suspension, the mixture was allowed to stand for 2 hr at 4°C and centrifuged at 2000 rpm for l0 min. After centrifugation, the supernatant containing phages was obtained, the phage suspensions were centrifuged at 16000 rpm for 2 h. at 4°C. Then, the supernatants were discarded and the pellets were resuspended in 1.0 ml of saline solution and then assayed.
2.9. Characterization of Bacillus thuringiensis Phages
- 2.9.1. Adsorption of UVAbsorption spectra of purified phages Bt1 and Bt2 were measured at serial wave lengths viz. 220, 240, 260, 280, 300 and 320 nm by 2 nm intervals in a spectrophotometer model UV vis Spectrophotometer UV 9100B, Lab Tech. in Biochemistry Dept., Faculty of Agriculture, Ain Shams Univ.2.9.2. Electron Microscopy ExaminationThe isolated phages were examined as described by Othman et al. [35]. A drop of the phage suspension (1012 pfu/ml) was placed on 200 mesh copper grids with carbon-coat formvar films, and the excess was drawn off with filter paper. A solution of 2% uranyl acetate was then placed on the grids and the excess drawn off as before. Specimens were examined using JOEL-JEM-1010 electron microscope (Electron Microscope Unite, Regional Center for Mycology and Biotechnology, Al-Azhar University, Cairo, Egypt).2.9.3. Lytic Pattern of the Bacillus PhagesLysosensibility of the isolated phages was carried out using the spot test technique as described by Sambrook et al. [36]. Plates containing base layers were inoculated with the over layer which containing the tested bacterial hosts (Bacillus subtilis, Bacillus thuringiensis, Bacillus polymyxa, Bacillus megaterium, Bacillus amyloliquefaciens, Bacillus lechiniformis and Bacillus circulans) separately in a 3 ml of melted semi-solid agar media, 20 µl of phage isolate was dropped on the surface of plates and then incubated for 24 hr 37ºC.2.9.4. Action of EcoRI Restriction Enzyme on B. thuringiensis Phages Genome2.9.4.1. Extraction of Phages DNAThe nucleic acid of the isolated phages was extracted according to the method described by Maniatis et al. [37] with minor modification by Compos et al. [38]. Fifteen ml of high titer phage suspension were incubated at 37°C for 30 min with DNase and RNase at final concentration of 1 µg/ml to get rid of contaminating bacterial DNA and RNA.Phage particles were precipitated by addition of NaCl and polyethylene glycol (PEG6000) to final concentrations of 3 and 5% (w/v), respectively. The mixture was incubated on ice for 30 min and centrifuged at 12,000 rpm for 20 min. The phage pellets were resuspended in 500 µl of 0.2 M ammonium acetate, 0.001 M Na-EDTA pH 7.5 and heated at 55°C for 20 min an equal volume of buffer saturated phenol was added and mixed by inverting the tube several times. The phases were separated by centrifugation at 13000 rpm for 5 min at room temperature in microcentrifuge. The aqueous phase was transferred to a clean tube and extracted once again with chloroform. The aqueous phase was recovered as described above and transferred to clean tube. The DNA was precipitated by adding 0.1 volume of 3 M sodium acetate pH 5.5 and equal vol. of cold ethanol followed by freezing for 7-8 hrs, the tube was thawed and DNA precipitate was collected by centrifugation at 13,000 rpm for 10 min. The pellet was washed in 70% ethanol, air dried at room temperature, resuspended gently in 30 µl of TE (10 mM tris-HCl pH 8.0, 1 mM EDTA) and stored at 4°C.2.9.4.2. Digestion of bacteriophages DNA with restriction enzymeThe reactions were assembled by mixing the components in a sterile 0.5 ml eppendorf (2 µl 10x buffer, 1 µg phage DNA, 3-5 units Restriction enzyme EcoRI and complete the total volume to 20 µl with Sterile deionized water). The reactions were incubated for 2 hrs at 37°C, and then 5 µl of bromophenol blue dye (450 mM tris borate pH 8.3, 50% (v/v) glycerol, 0.2% (w/v) bromophenol blue) were added and the reactions were heated at 60°C for 5 min to stop the reaction. The digested DNAs were stored at 4°C until ready for analysis by agarose gel electrophoresis. The digested DNA was separated in 1% agarose gel according to Peacock and Dingman [39] in the presence of 1 Kbp DNA ladder (biomol) as molecular weight standard to allow estimation of the DNA size of each bacteriophag. The stained gel was examined using an ultraviolet transilluminator with wavelength of 302 nm. The DNA bands which fluoresced orange were photographed using digital camera.
3. Results and Discussion
3.1. Enumeration of total Count of Bacteria
- The total count of bacteria were conducted by plate count technique using Tryptic soy agar medium. Results are shown in the following table (1):
|
3.2. Occurrence of Bacillus Phages Different Soils
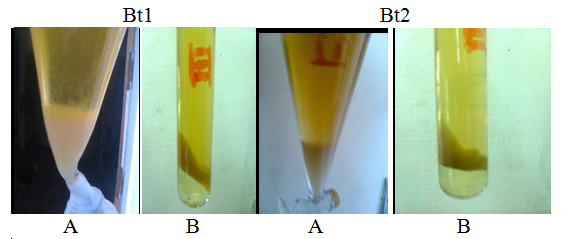
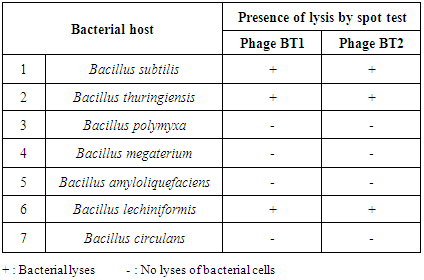
- Twelve different samples from the Egyptian deasert were tested using spot test technique (Fig. 1) for the presence of bacteriophages specific for Bacillus subtilis, Bacillus thuringiensis, Bacillus polymxa, Bacillus megaterium, Bacillus amyloliquefaciens, Bacillus lechiniformis and Bacillus circulans. The results (Table 2) revealed the presence of phages specific for Bacillus subtilis in samples 1 and 12 (Bahariya Oases and Great sand sea), phages specific for Bacillus thuringiensis occured in samples 1, 2 and 12 (Bahariya Oases, Mines and Great sand sea), phages specific for Bacillus polymxa at samples 1 and 11 (Bahariya Oases and Mountains negev 1), phages specific for Bacillus megaterium at samples 1 and 12 (Bahariya Oases and Great sand sea) and phages specific for Bacillus circulans at samples 9 and 10 (Abohrirh and Abumankar), while no phage was detected for Bacillus amyloliquefaciens and Bacillus lechiniformis at any soil sample as shown in table (2).
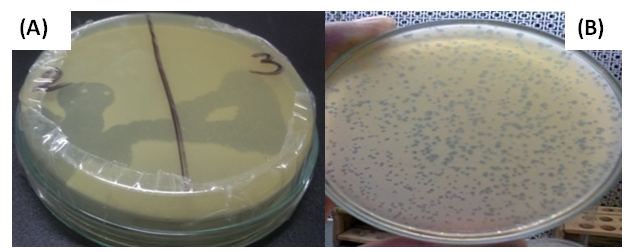 | Figure (1). Assaying of the phages specific for Bacillus qualitatively by the spot test (A) and quantitatively by the over layer technique (B) |
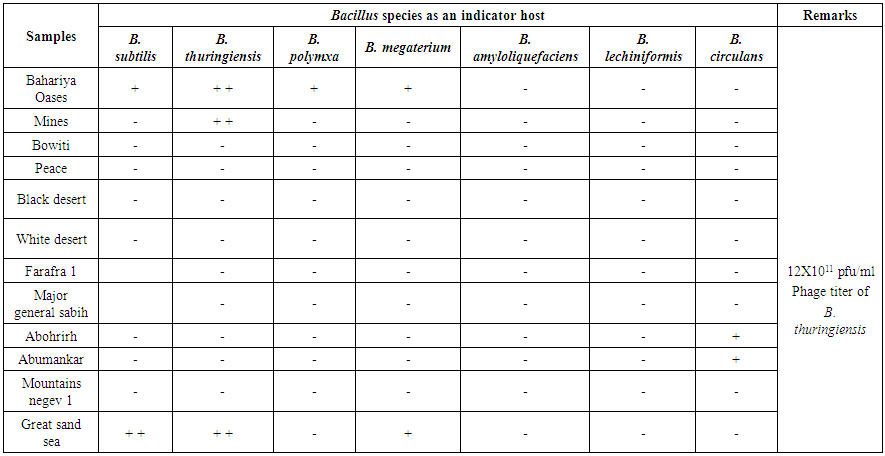 | Table (2). Detection of Bacillus phages in arid soils by the spot test technique |
3.3. Preparation of High Titer Phage Lysates
- High titer phage stocks were obtained using the liquid culture method as following: Erlenmeyer flasks (250 ml) containing 100 ml of liquid culture medium were prepared; each flask was inoculated with Bacillus thuringiensis and the phage lysate was added in a ratio of 1:10 and incubated at 30°C with shaking for 48 hr, after the incubation period, cultures were centrifuged at 6000 rpm for 15 min, then the suspension containing phages was assayed quantitavly and the phage count was 107, then transferred into sterile flasks and stored at 4°C with traces of chloroform.
3.4. Purification of Phages Specific for Bacillus thuringiensis
- Dextran sulfate-polyethylene glycol system (two phase system) was used [35] the weights 200, 14.2, 0.435 and 3.8 grams of phage lysate, polyethylene glycol (PEG 6000), dextran sulfate 500 and NaCl, respectively were mixed in a separating funnel (Fig 2.). After mixing the funnel was allowed to stand at 4°C overnight. A heavily turbid bottom layer was slowly collected into a clean tube and centrifuged at 2000 rpm for 15 min. The clear top and bottom phases were removed by pipette and the resulted remaining interface "cake" was 2.5 ml for phage 1 and 2 ml for phage 2, then they were suspended in 15 ml of a 1% (W/W) dextran sulfate solution. Then, 1.7 ml of a 3 M KCl was added to each milliliter of suspension; the mixture was allowed to stand for 2 hr at 4°C and centrifuged at 2000 rpm for l0 min.
3.5. Adsorption of UV
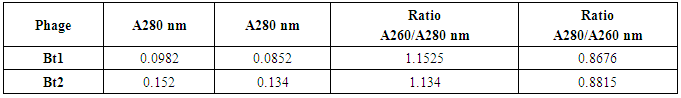
- Absorption spectra for purified viruses Bt1 and Bt2 phages were measured at serial wave lengths viz. 220, 240, 260, 280, 300 and 320 nm by 2 nm intervals. As shown in table (3), the purified phage particles of phages Bt1 and Bt2 suspended in saline solution have different absorption spectra at ultraviolet rays. Absorption of Bt1 phage at 260 nm was 0.0982, absorption at 280 nm was 0.0852, absorption at 260/280 nm was 1.1525, and absorption at 280/260 nm was 0.8676. Absorption of Bt2 phage at 260 nm was 0.152, absorption at 280 nm was 0.134, absorption at 260/280 nm was 1.134, and absorption at 280/260 nm was 0.8815.
|
3.6. Electron Microscopy Examination
- The particle morphology of the isolated viruses was determined by examination the negatively stained preparations using the transmission electron microscope. Electron micrograph (Fig. 3) shows that Bt1 phage has icosahedral capsid with hexagonal outlines with dimensions of about 140x165 nm and contractile tail type. The tail is about 117 nm length and 34 nm width, while the particle of phage Bt2 has long contractile tail with 206 nm length and 27 nm width. Its head Icosahedral capsid with hexagonal outlines with dimensions of about 140x124 nm.
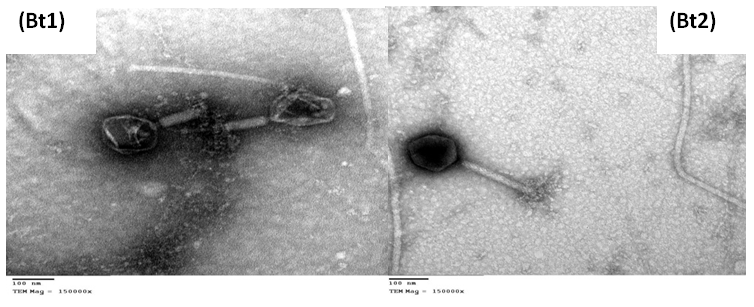 | Figure (3). Electron micrograph of purified Bacillus thuringiensis bacteriophages Bt1 and Bt2 negatively stained with 2% uranyl acetate using TEM (Magnification X-150000) |
3.7. Lytic Pattern of the Bacillus thuringiensis Phages
- Lysosensability of Bacillus subtilis, Bacillus thuringiensis, Bacillus polymyxa, Bacillus megaterium, Bacillus amyloliquefaciens, Bacillus lechiniformis and Bacillus circulans to the two isolated phages (specific for B. thuringiensis) was investigated qualitatively by spot test technique. Data represented in table (4) show that, B. thuringiensis phage Bt1 and phage Bt2 both have a narrow host range and able to lyses the main host Bacillus thuringiensis and another two bacterial hosts Bacillus subtilis and Bacillus lechiniformis. El-Sayed et al. [42] isolated phages specific to Bacillus cereus and Bacillus thuringiensis strains and mentioned that the phages have narrow host range.
|
3.8. Digestion of Bacteriophages DNA with Restriction Enzyme
- The Bt1 and Bt2 phages had a linear double-stranded (ds) DNA because they were susceptible to digestion with restriction enzyme endonucleases, EcoRI and subsequently subjected to electrophoretic analyses. As shown in Fig (4), the phage DNA was sensitive to EcoRI. The genomes were fragmented into four fragments with size of 1500, 1400, 800 and 510 bp. They were estimated to be 4.210 kbp for Bt1 phage, as determined by combining the fragment sizes obtained with the restriction enzyme used, and 6.480 kbp for Bt2 phage with 4 fragments with size of 3000, 1600, 1350 and 530 pb.
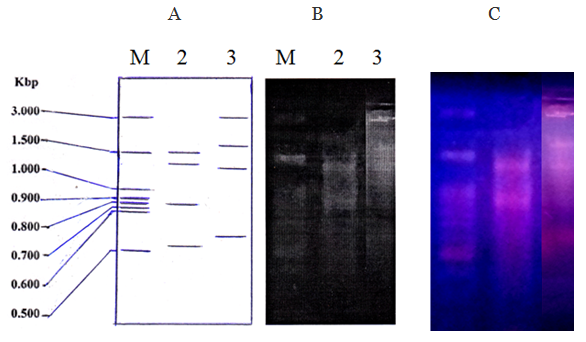 | Figure (4). A diagram (A) and a photograph (B) of DNA restriction patterns of bacteriophages Bt1 and Bt2 digested with EcoRI. (C) The gel under ultraviolet transilluminator. M= Marker DNA |
4. Conclusions
- In this study two phages specific for Bacillus thuringiensis were isolated from arid soil in Egypt, they were assayed qualitatively and quantitatively, propagated on their specific bacteria, purified, morphologicaly characterized, their host range was studied, and the action of EcoR1 restriction enzyme on their genomes was studied.
ACKNOWLEDGMENTS
- I would like to express my deep and sincere gratitude to Prof. Dr. Badawi A. Othman; Virology lab., Agricultural Microbiology Dept., Faculty of Agriculture, Ain Shams University, Cairo, Egypt, for his kindly provided the different identified bacterial isolates and other equipment. Special appreciation and thanks goes to Prof. Dr. Adel M. Hammad; Agricultural Microbiology Dept., Faculty of Agriculture, El-Menia University, El-Menia, Egypt for his experimental support during the digestion of bacteriophages DNA with restriction enzyme.
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML