-
Paper Information
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
International Journal of Virology and Molecular Biology
p-ISSN: 2163-2219 e-ISSN: 2163-2227
2016; 5(1): 16-26
doi:10.5923/j.ijvmb.20160501.03

Comparative Sequence Analysis of Different Strains of African Swine Fever Virus Outer Proteins Encoding Genes from Nigeria, 2009 – 2014
Pam D. Luka 1, 2, Joseph Erume 2, Frank N. Mwiine 2, David Shamaki 3, Bitrus Yakubu 1
1Biotechnology Division, National Veterinary Research Institute, Vom, Nigeria
2College of Veterinary Medicine, Animal Resources and Biosecurity, Makerere University, Kampala, Uganda
3Directorate of Research, National Veterinary Research Institute, Vom, Nigeria
Correspondence to: Pam D. Luka , Biotechnology Division, National Veterinary Research Institute, Vom, Nigeria.
| Email: |  |
Copyright © 2016 Scientific & Academic Publishing. All Rights Reserved.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/

African swine fever (ASF) is an economically important disease of domestic pigs causing a huge amount of losses. Understanding the extent and dynamics of genetic diversity of genes coating outer proteins is required for a rational vaccine design and interpretation of efficacy of vaccine or therapy for the control of ASF. This study was designed to investigate the nucleotide structure of genes: E183L, KP177R and O61R encoding outer proteins p54, p22 and p12, respectively, of African swine fever virus isolates from Nigeria for genetic variability. The samples were collected over three years and analyzed for relatedness using MEGA5 and Hopp and Woods procedure for predicting protein antigenic determinants. The result of our comparative study revealed variability in p12 sequences of the isolates collected from different locations within the country. The p54 and p22 genes were observed to be more conserved among the isolates. Hydropathy analysis of all the genes failed to reveal any structural variability within the proteins of interest. Our study revealed mutations (insertions/deletions) within the 3’ terminus of the p12 gene thus we conclude that p12 is under selection pressure and therefore, its utility needs to be further assessed widely for genetic diversity, antigenicity and pathogenicity of the virus.
Keywords: African swine fever virus, p54 gene, p22 gene, p12 gene, Comparative sequence analysis, Hydropathy profile, Nigeria
Cite this paper: Pam D. Luka , Joseph Erume , Frank N. Mwiine , David Shamaki , Bitrus Yakubu , Comparative Sequence Analysis of Different Strains of African Swine Fever Virus Outer Proteins Encoding Genes from Nigeria, 2009 – 2014, International Journal of Virology and Molecular Biology, Vol. 5 No. 1, 2016, pp. 16-26. doi: 10.5923/j.ijvmb.20160501.03.
Article Outline
1. Introduction
- African swine fever (ASF) is an economically significant disease of pigs caused by a large DNA virus and a sole member of the family Asfarviridae and genus Asfivirus 1 [1]. African swine fever virus (ASFV) within an infected host, replicates in the cytoplasm with variable levels of virulence that ranges from highly lethal to subclinical infections. The virus within an infected host elicits a peculiar immune response in which no neutralizing antibodies produced are effective and apparently healthy animals become carriers [2]. Both domestic and wild pigs are susceptible to the virus with the soft tick (Ornithodoros genus) being the primary reservoir. Wild African suids such as warthogs and bush pigs have been reported to be infected but hitherto remained clinically asymptomatic. The ASF virion is ~200 nm in diameter and contains more than 50 proteins and consists of several concentric layers enclosing an electron-dense nucleoid, containing a double stranded DNA genome of 170-190 kilobase pairs (kbp). The viral core is enwrapped by an inner lipid envelope beneath the icosahedral capsid [3]. The extracellular particles also possess an additional envelope which is derived from the plasma membrane. However, on analysis of cell extracts at various life cycle phases of the virus revealed more than 100 viral proteins [4].ASF has been reported in most sub-Saharan Africa where the virus is maintained within a sylvatic cycle that involves soft tick (Ornithodoros genus) infecting wild pigs with asymptomatic effects. The virus then replicates in the tick before being transmitted to wild swine through blood meal and the cycle maintained indefinitely. Interestingly, these has facilitated the persistence and emergence of new variants in east, central and southern Africa. However, a domestic cycle with or without the involvement of ticks also occur and common in West Africa [5].The eponymous disease was first described in Kenya 1921 and later thought to be native to the African continent. However, from the first report in 1920s, the virus made an inroad into Europe with devastating effects in Spain, France and Belgium in 1957 and 1960s before being eradicated [6]. Eventually the disease became endemic in the island of Sardinia [7]. Recently, the disease was re-introduced into the borders of Europe with Ukraine, Georgia, Poland, Ukraine, the Caucasus and the Russian federation being affected and posing varying risk of introduction to other European ASF free countries [8-10].Several factors such as genome length variation between different ASFV isolates, host susceptibility, presence of tick vectors, and the probable interaction amongst host, suids and vectors has been associated with virulence and pathogenicity of viral isolates [11, 12]. Structurally, the ASFV p54 protein is a viral structural protein of 25-27 kDa, which is externally located. It’s a putative transmembrane domain localized at the endoplasmic reticulum (ER)-derived envelope precursors. The protein is encoded by the E183L gene open reading frame (ORF) [13] and critical for the recruitment and transformation of the ER membranes into the precursors of the viral envelope for the virus viability and early viral infection and adsorption to susceptible host cells [14]. Other published reports have also shown that p12, p22, and p30 are equally essential viral proteins that are also involved in early events of replication [15].The p22 protein is encoded by the gene - ORF KP177R, has an apparent molecular weight 22 kDa and located externally in the viral particle and carries a hydrophobic domain that is characterized by a signal peptide. Whereas the ASFV virus protein p12 (ORF O61R) mediates in the adsorption of ASFV to susceptible cells via the structural virus proteins located within the outer envelope of the virion. The protein appears and accumulates late after the earlier phases of infection have occurred without undergoing posttranslational modification to become functional [13, 16, 17]. Previous studies reported high level of conservation of the 5’ flanking region of the p12 gene and high variability of the downstream 3’ end [18]. However, Vlasova et al., [19] in a comparative study of the three genes revealed that the p12 gene seems to be under selection pressure.Since there is no available vaccine against ASF, stringent bio-security and rapid diagnostic response are options for the control and eradication of the disease in sub-Saharan Africa and Nigeria. Nevertheless, several vaccine studies have been conducted but with minimal or no success of obtaining long-term neutralizing antibodies. This lack of ASFV vaccines might be due to unique molecular and biological properties of the virus and the interactions of its proteins which are responsible for virus–cell interactions [20, 21].The virus within infected host elicits certain immune responses from its structural proteins to which neutralizing antibodies are produced against them [15, 20]. Extensive studies into the genetic structure, genes, and immune mechanisms of the virus which affect viral replication, virus–host interactions, and virulence will help in understanding the viral biological characteristics and mechanism of disease for comprehensive and effective control strategies for the agent to be achieved [6, 22].During infection, the host cell comes in contact with both external and proteins structures of the virus. This interaction stimulates the production of immune structures for effective control. As a result, the aim of this study was to investigate the nucleotide structure of genes encoding outer proteins p54, p22 andp12 of African swine fever virus isolates from Nigeria and compare with others obtained globally for possible variation.
2. Materials and Methods
2.1. Study Area and Samples
- Nigeria is a federated country comprising 36 states and the Federal Capital Territory and bordered by four countries namely Benin, Niger, Chad, Cameroon and the Atlantic Ocean. Pig production is concentrated in the south western, central and eastern parts of the country but restricted in the north due to cultural and religious bias. In this study, 31 ASF confirmed positive samples collected by field veterinarians and submitted to the Central Diagnostics Laboratory (CDL) of the National Veterinary Research Institute (NVRI), Vom Nigeria from 2009 – 2014 were used for this study. The samples were collected from domestic pigs from 6 states (Anambra, Benue, Cross River, Delta, Lagos and Plateau states).
2.2. DNA Amplification of ASFV p12, p22 and p54 genes
- DNA from field samples were extracted from liver, spleen, and mediasternal lymph nodes using QIAamp DNA mini kit (Qiagen, Hilden, Germany) according to the manufacturer’s specification. Lyophilized freeze-dried E70 from the European Union reference laboratory for ASF (CISA-INIA, Madrid Spain) was used as control for this study. The expected genes were amplified with primers according to protocols earlier described by Nielan et al., [20] and Angulo et al., [18]. The PCR products were resolved on 2% agarose gel.
2.3. DNA Sequencing and Sequence Analysis
- Amplicons obtained were purified using the Wizard® SV Gel and PCR Clean-Up System (Promega, Madison USA) according to manufacturers’ specifications and shipped to Inqaba biotec®, South Africa for sequencing. The three gene segments (p54, p22 and p12) were sequenced using the ABI Big Dye V3.1 kit cycle sequencing on the ABI PRISM® 3100 Genetic Analyzer (Applied Biosystems).Complete alignment of nucleotide sequences was performed using ClustalW. The neighbour-joining method in MEGA version 5 was used with 1,000 bootstrap replications to determine phylogenetic relationship. Sequences obtained were comparatively analysed with those retrieved from the GenBank. Deduced amino acids (a.a.) was determined by BioEdit software version 7.2.5. The sequence of Georgia 2007/1 strain complete genome (Accession number FR682468.1) was used for analyses and for comparison as reference sequences of genes p54 (protein ID CBW46791.1), p22 (protein ID CBW46645.1), p12 (protein ID CBW46764.1).
2.4. Hydropathy Profiles Analysis
- The initial step in search of antigenic determinants [24] in pathogenic organism is by analysing the protein(s) that antibody binds. Therefore, deduced amino acids were analysed for antigenic determinants. Hydrophobic amino acids on protein folding and function were also analysed based on Hoop and Wood [25] method on www.web.expasy.org/protscale/
3. Result
3.1. Sequence Analysis
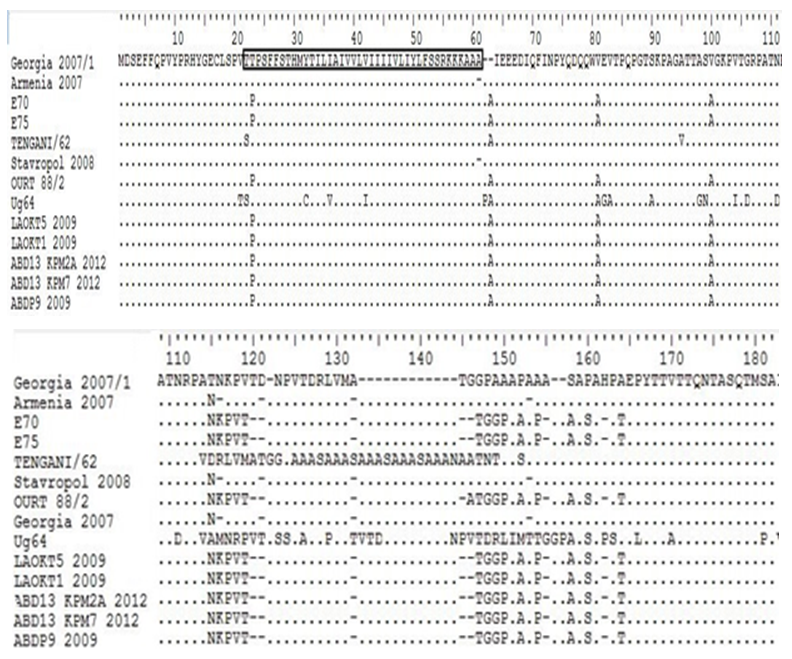
- The expected 603, 534 and 138 bp fragments for the corresponding p54, p22 and p12 genes respectively were amplified from the 31 ASF positive tissue samples collected. All the positive samples amplified a single and discrete corresponding amplicon. The nucleotide sequences of the ASFV isolates were deposited into GenBank (accession nos. p54: KT150945-150973 KT961344-961363; p22: KU641719-641749 and p12: KU641698-641718).Multiple sequence alignments were done to eliminate misalignments and ensure that the sequence coding frame are correct. All gaps were considered as missing information used to avoid artificial nucleotide divergence in the phylogeny. However, none of the different methods used in RDP3 package identified recombination events in E183L and KP177R sequences. Several recombination events were observed in O61R (data not shown).Sequence analysis revealed that all the nucleotide sequences of the gene encoding p54 from Nigeria were identical to each other and to Espana70 and E75 sequences (Figure 1, only four sequences are shown). Between positions 180-190, all the Russian isolates have 6 nt deletions while Nigeria and European have 3nt. The Ug64 isolate do not have a deletion at that position. Also at positions 458, 6nt deletions were also observed for both Nigerian, European and Russian isolates with the exception of Tengani62 and Ug64 isolates.
 | Figure 1. Multiple sequence alignment of p54 gene sequences of ASFV isolates where: (points) denotes identical nucleotide, - (dash) indicates deletions of nucleotide |
 | Figure 2. Multiple sequence alignment of p22 gene sequences of ASFV isolates where: (points) denotes identical nucleotide, - (dash) indicates deletions of nucleotide |
 | Figure 3. Multiple sequence alignment of p12 gene sequences of ASFV isolates where:. (points) denotes identical nucleotide, - (dash) indicates deletions of nucleotide |
3.2. Hydropathy and Amino Acid Analysis
- All the genes analyzed (p54, p22 and p12) showed single nucleotide substitutions. The substitution within p12 is within the 3’end (N-terminal) which has a net positive charge but not present in Nigerian and European sequences. However, analysis of these changes on the functional role of encoded proteins carried out revealed similar outcome as reported by Vlasova et al., [19]. Deduced amino acid sequence analysis and hydrophilicity profile of p54 revealed a 32aa stretch of hydrophobic residues and a 130aa long hydrophilic c-terminal. The N-terminal of all p54 sequences are conserved however, using reference sequence (Georgia 2007/1) a number of mutations were observed i.e. threonine to proline in African and European (E70, E75) strains but non on the eastern European strains. Only the Ugandan strain (Ug64) showed 3 mutations of tyrosine to Cysteine, Isoleucine to valine, and valine to isoleucine within the hydrophobic residues. Mutations to alanine was more prevalent with a highly variable c-terminal (Figure 4).
3.3. Phylogenetic Analysis of ASFV Isolates
- The results of phylogenetic analysis of the three (p54, p22, p12) genes obtained from 31 ASFVs from Nigeria are presented. A total of 26 sequences (p54) obtained from this study were compared with sequences from the GenBank database to infer phylogenetic relationship using neighbor-joining. The Nigerian p54 cluster with other European isolates, the other clusters are from the Armenia and Russian Federation and the southern Africa isolates (Tengani and Ug64) (Figure 7).
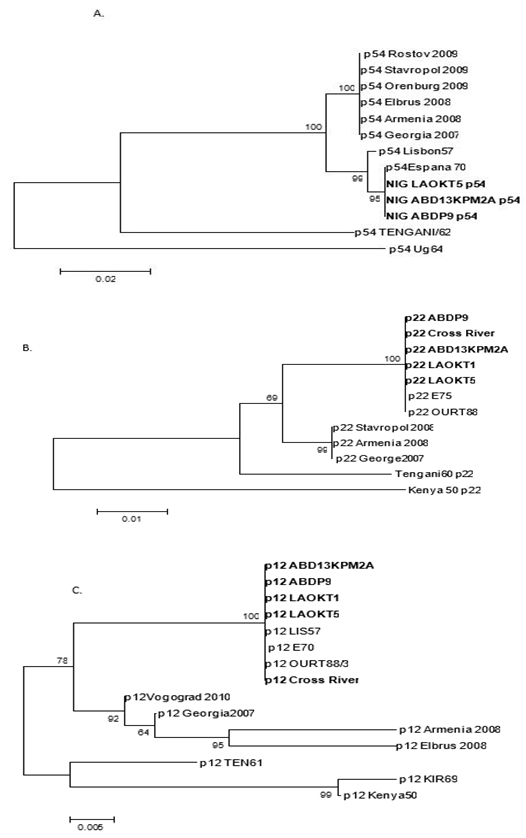 | Figure 7. Phylogenetic relationship of E183L (p54) genes (A), KP177R (p22) gene (B) and O61R (p12) gene (C) sequences of some representatives |
4. Discussion
- ASF is a major economic and transboundary disease of pigs that causes significant mortality. Morbidity and mortality have continued to vary due to varying virulence of the virus within an infected area. Several authors had described high levels of variabilities across discrete regions which have been useful in differentiating closely related strains of the virus. However, none of the structural heterogeneity is up to 40% [18, 26].Within the framework of Hoop and Woods [25] procedure, hydropathy profile has been reported to predict and or corresponds to adjacent antigenic determinants in relation to the functional properties of the protein structure. The ASFV deduced amino acids proteins sequences from our study were generally conserved and similar to previous results obtained by other workers [18, 19, 27] but with minor variations (silent or conversed mutations) which translate into a changes such as Isoleucine to valine, glycine to serine glutamic acid to aspartic acid as observed in p12. This gene encoding an outer protein have been reported to be under selection pressure by their involvement in attachment and penetration into the host cell. Our finding is in agreement with Angulo et al [18] who suggested that conservation of the p12 polypeptide sequence was associated with selective pressure against mutations that alters the basic properties of the polypeptide due to its essential role in infection. Additionally, the p12 protein putative transmembrane domain that anchor the polypeptide chain externally and a cysteine-rich domain in the C-terminal region was suggested to account for multimerization through a disulphide bond without posttranslational modification [16]. Interestingly, these mutations can influence antigenicity.The p22 (KP177R) proteins are localised on viral envelope as a single copy gene in BA71V and Georgia 2007/1 [33]. Our findings agree with Chapman et al [34] who reported that the p22 protein in Benin 97/1 and OURT 88/3 are 100% conserved. Besides, some genomes have been reported to encode 2 copies of the p22-related ORF which are quite divergent and expression of these proteins has been suggested to contribute to antigenic variation of the virus [33].ASFV p54 is one of the most variable glycoprotein. Our study observed high level of variability across the amino acid sequence with the C-terminal showing the most variability. This is in agreement with the findings of Mima et al [35] who demonstrated mostly variability on the C-terminal region which is localized on the inner side of the cell membrane. However, high level of differences in the nucleotide sequences of p54gene (E183L) for various ASFV isolates used in this study may be the result of random mutations during virus evolution. Genotically, several gene segments such as p72, p54, and central variable region (CVR) have being used to characterize ASF virus. Several genotypes have been resolved globally with a majority of the genotypes circulating in East and Southern African due to the presence of wild pigs and tick vector [28]. Sequence analysis also suggests that the circulating viruses in Nigeria are homogenous and therefore reaffirms genotype I as the only circulating genotype in Nigeria [29, 30].A phylogenetic analysis showed that all the Nigerian sequences cluster together separately from European, Russian/Eastern Europe and East/Southern Africa. Our findings suggest that the sequences are homogenous at the regional level and heterogeneous globally. This is in agreement with Vlasova et al [19] who reported a similar clustering of isolates from the Russian Federation and Armenia differently from sequences from Europe and Africa.The p12 phylogenetic analysis also followed a similar pattern with those reported by Vlasova et al., [19] where the Nigerian, Russian, European, and Southern African strains cluster differently from each other. All the p12 sequences were conserved at the 5’ terminus whereas high level of variability at the 3’ terminus was observed. This is in agreement with findings of Angulo et al., [18]. The role of these variations in adaptation or virulence is yet to be determined. Nevertheless, based on Michaud et al., [31] and Rowlands et al., [32], the Russian cluster belong to genotype II, Nigeria and Europe genotype I, Ten61 genotype V, KIR69 and Kenya1950 genotype X.
5. Conclusions
- There is scanty information in the GenBank for p12 sequences but for a wider comparison however, sequence analysis of the Nigerian and European strains observed deletions at the 3’flanking terminus of p12 and insertions for the Russian and Southern African strain. Our findings revealed that the p12 gene is more prone to mutation thus we conclude that p12 might be prone to selection pressure and its utility needs to be further assessed widely for genetic diversity. Although there is minimal diversity among p54 and p22 genes, it would be ideal to have a good representation of the diversity of circulating ASFV structural protein genes worldwide when thinking of vaccine design and efficacy. Considering that selective pressure plays an important role in structuring the diversity of antigens however, a single strain or gene may not represent global circulating strains if there is synergy between two or more genes.
ACKNOWLEDGEMENTS
- We are grateful to the technical staff of Biotechnology Division of the National Veterinary Research Institute and the Federal department of Veterinary and pest control services of the Federal Ministry of Agriculture and Rural development, Abuja. This study was partly sponsored by National Veterinary Research Institute, Vom, Plateau State, Nigeria, and the IAEA Research Contract No: 18347/R0.
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML